Pick Up
787. Genome Editing Produces Rice Plants with Improved Yield under Phosphorus Deficiency
787. Genome Editing Produces Rice Plants with Improved Yield under Phosphorus Deficiency
Tillering is the branching of rice plants and is an important trait that determines shoot architecture and yield. The number of tillers is determined by the genes of each rice cultivar and environmental factors. Many genes are involved in determining the number of tillers, and among them, rice TEOSINTE BRANCHED1 (OsTB1) is known to be an important gene that suppresses tillering. On the other hand, phosphorus, a soil nutrient, is one of the most important environmental factors involved in tillering. Rice plants grown under phosphorus deficiency have a greatly reduced number of tillers, which is a major constraint for rice production in sub-Saharan Africa, where phosphorus-deficient soils are widespread.
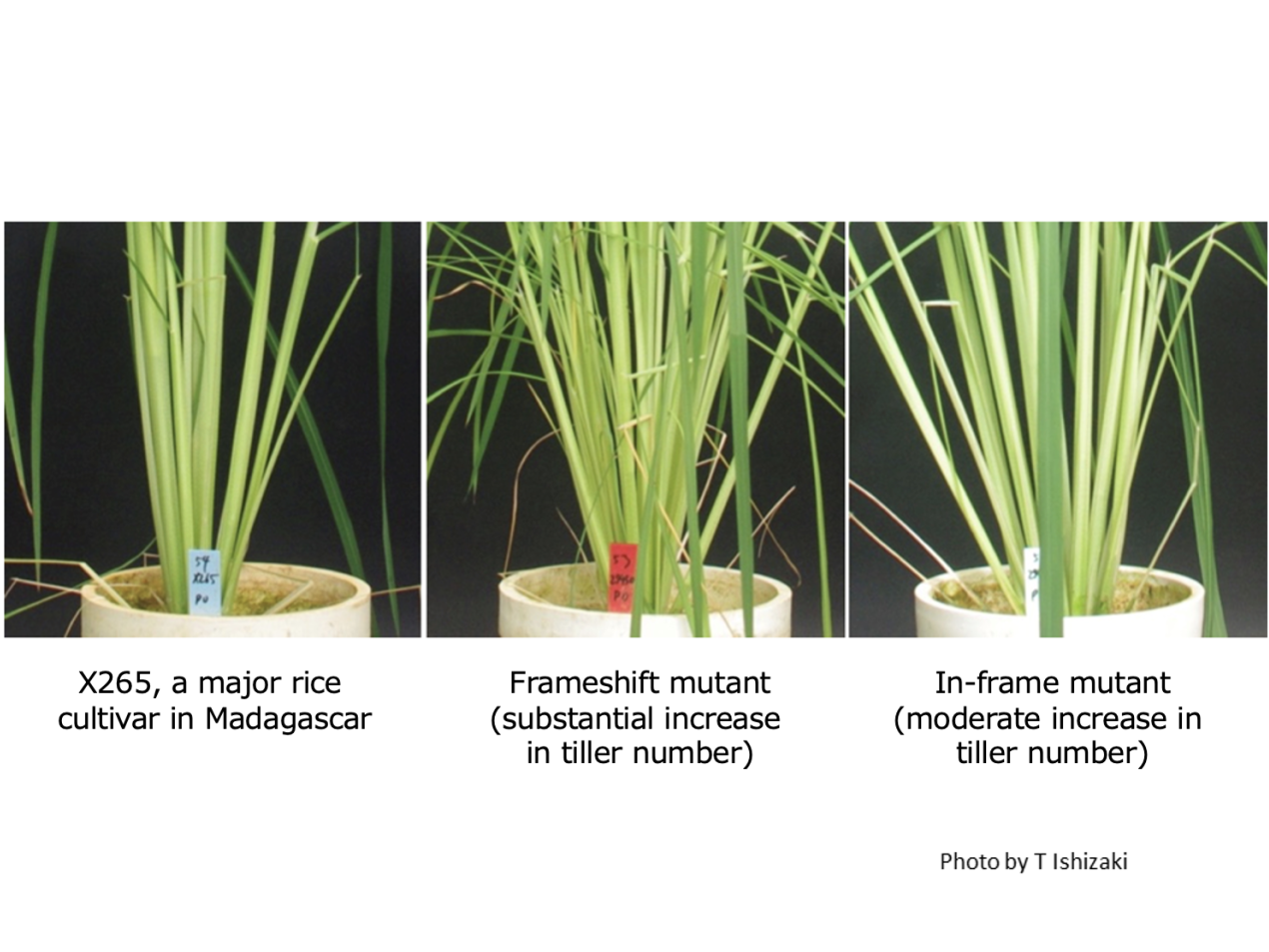
Using the CRISPR/Cas9 system, a genome editing technique, we have generated two types of mutant lines for the OsTB1 gene of X265, a major rice cultivar in Madagascar (Ishizaki et al. 2023). One is a frameshift mutant (single base insertion) and the other is an in-frame mutant (30-base deletion) (see Note). When grown in nutrient-rich soil, the original X265 had about 18 tillers, the frameshift mutant line had about 62 tillers, and the in-frame mutant line had about 22 tillers at the maximum tillering stage. This means that the OsTB1 gene loses its tillering suppression function through the frameshift mutation, while the in-frame mutation weakens its tillering suppression function. When these mutant lines were grown under low-phosphorus condition, the frameshift mutant line increased the number of tillers by about 5-fold compared to X265, but did not increase yield due to a significant reduction in thousand-grain weight and a lower filled grain rate. On the other hand, the in-frame mutant had about 1.4 times the number of tillers of X265, and although a slight reduction in thousand-grain weight was observed, the yield was increased by 38% with the same filled grain rate as that of X265. Thus, we have shown that a moderate increase in the number of tillers due to the in-frame mutation of OsTB1 can contribute to yield improvement under phosphorus deficiency.
Recently, a joint research group from JIRCAS, the National Agriculture and Food Research Organization, Nagoya University, Yokohama City University, RIKEN, Meiji University, and Kazusa DNA Research Institute identified MP3 (MORE PANICLES 3), a gene derived from Koshihikari rice cultivar that increases yield under high atmospheric CO2 level (Takai et al. 2023). MP3 is an allele of the OsTB1 gene and, like in-frame mutants created by genome editing, has the effect of moderately increasing the number of tillers. We believe that OsTB1 with the in-frame mutation and MP3 genes have the potential to improve rice productivity in areas with poor fertilizer and soil phosphorus supply, such as sub-Saharan Africa, and also to increase rice yield in high CO2 environments.
Note: The DNA sequence is commonly referred to as the nucleotide sequence, and three nucleotides correspond to one amino acid. A frameshift mutation is one in which the reading frame of the amino acid code is shifted by the insertion or deletion of nucleotides that are not multiples of three, resulting in an abnormal protein with a completely different amino acid sequence. On the other hand, in-frame mutations, where the number of nucleotides involved in the mutation is a multiple of three, do not shift the reading frame of the amino acid coding triplet, so that a protein with the same amino acid sequence as the original protein is produced in the unmutated region.
References
Ishizaki, T., Ueda, Y., Takai, T., Maruyama, K., Tsujimoto, Y. (2023) In-frame mutation in rice TEOSINTE BRANCHED1 (OsTB1) improves productivity under phosphorus deficiency. Plant Science, https://doi.org/10.1016/j.plantsci.2023.111627
Takai, T., Taniguchi, Y., Takahashi, M., Nagasaki, H., Yamamoto, E., Hirose, S., Hara, N., Akashi, H., Ito, J., Arai-Sanoh, Y., Hori, K., Fukuoka, S., Sakai, H., Tokida, T., Usui, Y., Nakamura, H., Kawamura, K., Asai, H., Ishizaki, T., Maruyama, K., Mochida, K., Kobayashi, N., Kondo, M., Tsuji, H., Tsujimoto, Y., Hasegawa, T., Uga, Y. (2023) MORE PANICLES 3, a natural allele of OsTB1/FC1, impacts rice yield in paddy fields at elevated CO2 levels. The Plant Journal, https://doi.org/10.1111/tpj.16143
Discovery of Koshihikari-Derived Gene that Increases Rice Yield in High CO2 Environment―Contributing to sustainable rice cultivation under climate change―https://www.jircas.go.jp/ja/release/2022/press202225
Contributors: ISHIZAKI Takuma (TARF); TSUJIMOTO Yasuhiro, TAKAI Toshiyuki and UEDA Yoshiaki (Crop, Livestock and Environment Division); MARUYAMA Kyonoshin (Biological Resources and Post-harvest Division); NAKASHIMA Kazuo (Food Program)
