Genome editing systems in rice cultivars and strategy for producing desired mutants with homozygous mutation
Description
CRISPR/Cas9 is a novel tool for targeted mutagenesis and is applicable to plants, including rice. The reported studies used limited rice cultivars that have high transformation efficiencies but are now used only for research purposes. For practical application of genome editing to molecular breeding in rice, there is a need to establish CRISPR/Cas9 systems that can be used in commercial cultivars. In targeted mutagenesis, biallelic homozygous mutants in which the target mutations are inherited stably by later generations are desirable for molecular breeding. Previous reports on CRISPR/Cas9 in rice have demonstrated that target mutations are transmitted to the next generation in accordance with Mendelian law, but heritability of the target mutation and the role of inherited Cas9 gene have not been fully elucidated. Here, we targeted the rice phytoene desaturase (OsPDS), whose mutants exhibit an albino phenotype, in five rice cultivars by using CRISPR/Cas9 and analyzed the segregation of target mutations. We present a strategy for generating homozygous mutants without transgenes, chimerism, or unpredicted mutations by using CRISPR/Cas9 in rice.
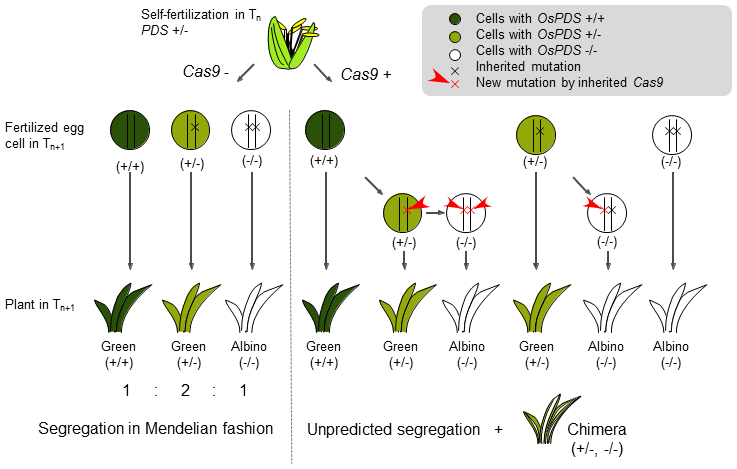
Agrobacterium-mediated methods using immature embryos successfully transformed a CRISPR/Cas9 system into five rice cultivars including commercially important ones and subsequently induced mutation (Table 1). Unpredicted segregations, with more mutants than theoretically predicted, were frequently found in T1 plants from monoallelic T0 mutants. Chimeric plants with both biallelic and monoallelic mutated cells were also observed in the T1 (Fig. 1). Next, we followed the segregation of a target mutation in the T2 from monoallelic T1 mutants. When T1 mutants possessed Cas9, unpredicted segregations of the target mutation and chimeric plants were observed again in the T2. When T1 mutants did not possess Cas9, segregation of the target mutations followed Mendelian law and no chimeric plants appeared in the T2. T2 mutants with Cas9 had mutations different from the original ones found in T0. Our results indicated that inherited Cas9 was still active in later generations and could induce new mutations in the progeny, leading to chimerism and unpredicted segregation (Fig. 2). We conclude that Cas9 must be eliminated by segregation in T1 to generate homozygous mutants without chimerism or unpredicted segregation.
Genes of commercially important rice cultivars can be modified by using CRISPR/Cas9 through our transformation systems, as has been done for other “model” cultivars. Investigators also must consider the possibility of unpredicted segregation caused by somatic mutation in T0, off-target mutagenesis, and somaclonal variation, which usually can occur in CRISPR/Cas9 technologies. For the practical use of mutants produced by our systems, intellectual property rights must be handled accordingly.
Figure, table
-
Table 1. Genotypes of rice cultivar mutants of OsPDS, produced by using the CRISPR/Cas9 system and by Agrobacterium-mediated transformation of immature embryos.
CultivarNumber of transgenic plantsNumber of monoallelic mutantsNumber of biallelic mutantsTotal number of mutantsNipponbare106266490Koshihikari1711011NERICA154163349Curinga27213780217IR645415Total454184188372 -
Fig. 1. Segregation of chimeric plants in the T1.
(a) Leaf of chimeric plant at seedling stage. (b) Leaves of chimeric plants grown in a greenhouse. (c) Spikelet of a chimeric plant. Bars represent 1 cm. (d) Green and white portions of chimeric plants were separately subjected to PCR-RE assay for OsPDS and to PCR assay for Cas9. In the case of monoallelic mutants, PCR-RE assay indicated three bands: 718 bp, 320 bp and 157 bp. In the case of biallelic mutants, PCR-RE assay indicated only a single band: 718 bp. Expected size of PCR products of Cas9 was 320 bp. -
Fig. 2. Diagrammatic model of unpredicted segregation. When plants do not inherit Cas9, the targeted mutation segregates in a Mendelian fashion. When plants inherit Cas9, the Cas9 can induce new mutations leading to unpredicted segregation and chimerism.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Tropical Agriculture Research Front
- Research project
- Program name
- Term of research
-
FY2016(FY2014~FY2016)
- Responsible researcher
-
Ishizaki Takuma ( Tropical Agriculture Research Front )
- ほか
- Publication, etc.
-
https://doi.org/10.1007/s11032-016-0591-7
Ishizaki T (2016) Molecular Breeding, 36
- Japanese PDF
-
A4 381.14 KB
A3 311.62 KB
- English PDF
-
A4 664.3 KB
A3 619.81 KB
- Poster PDF
-
Poster 419.24 KB