Functional analysis of genes in quinoa using a virus vector system
Description
Quinoa (Chenopodium quinoa Willd.) is an annual protein-rich pseudocereal native to the Andean region of South America. Quinoa has been recognized as a potentially important crop in terms of global food and nutrition security since it can thrive in harsh environments and has an excellent nutritional profile. JIRCAS and collaborative researchers have been analyzing the complex and heterogeneous allotetraploid genome of quinoa, and have recently overcome the challenges, with the whole genome-sequencing of quinoa and the creation of genotyped inbred lines (Research Highlights 2016, B03: Draft genome sequence of an inbred line of Chenopodium quinoa, an allotetraploid pseudocereal crop with high nutritional properties and tolerance to abiotic stresses; Research Highlights 2020, B08: Genetic and phenotypic variation of agronomic traits and salt tolerance among quinoa inbred lines). However, the lack of technology to analyze gene function in planta is a major limiting factor in quinoa research.
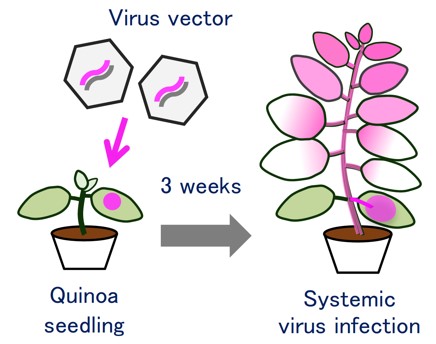
In this study, we demonstrate that the virus-mediated transient gene expression or repression techniques can be used in quinoa plants (Fig. 1). We show that apple latent spherical virus (ALSV) vector can induce gene silencing of a quinoa carotenoid biosynthesis gene, phytoene desaturase (CqPDS1) (Fig. 2). Virus-mediated silencing of CqPDS1 induces decreased accumulation of carotenoids and causes photobleaching symptoms in quinoa plants (Fig. 3). We also show that ALSV-mediated gene silencing can also be used in a broad range of quinoa inbred lines derived from the northern and southern highland and lowland sub-populations. Our data also indicate that repression of a quinoa 3,4-dihydroxyphenylalanine 4,5-dioxygenase gene (CqDODA1) or a cytochrome P450 enzyme gene (CqCYP76AD1) reduces accumulation of red-violet betalain pigments in quinoa plants (Fig. 4).
Our data demonstrate that the virus vector system is a useful tool for evaluating gene function in quinoa, where molecular breeding techniques such as genetic transformation have not been developed yet. Functional validation of quinoa genes, utilizing the published genomic information, could provide gene resources for molecular breeding of quinoa.
Figure, table
-
Fig. 1. Schematic of the virus vector system in quinoa plants
-
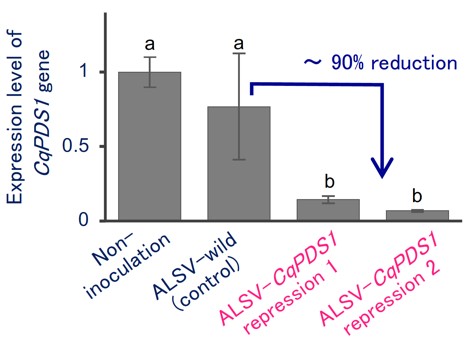
Fig. 2. ALSV induces silencing of carotenoid biosynthesis genes CqPDS1 in quinoa
RT-qPCR quantification of CqPDS1 transcripts in the uninoculated upper leaves of plants inoculated with the indicated inocula. Data are normalized and are shown as means ± SD (n = 3). Different letters indicate significant differences (p < 0.05). -
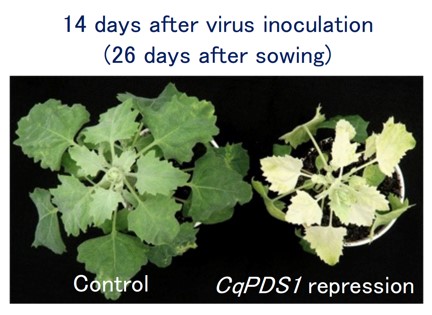
Fig. 3. Silencing of CqPDS1 induces photobleaching phenotypes in quinoa plants
A representative image of quinoa plants (inbred Iw line) at 14 days after virus inoculation with ALSV-wild (control) and ALSV-CqPDS1 is shown. -
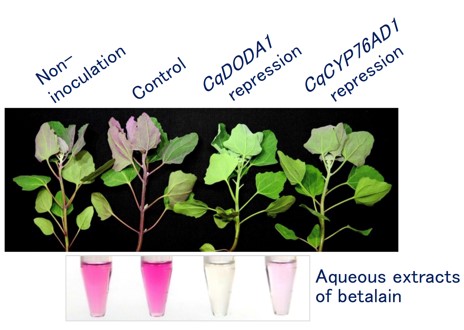
Fig. 4. Functional analysis of genes for betalain pigments biosynthesis in quinoa using the virus vector system
Virus-mediated silencing of CqDODA1 and CqCYP76AD1 inhibits betalain production in quinoa. Representative images of quinoa plants (inbred J056 lines) and aqueous extracts from the uninoculated upper leaves are shown. Quinoa plants inoculated with ALSV-wild were used as a control.
Figures reprinted/modified with permission from Ogata et al. (2021).
- Classification
-
Research
- Research project
- Program name
- Term of research
-
FY 2016–2025
- Responsible researcher
-
Ogata Takuya ( Biological Resources and Post-harvest Division )
Toyoshima Masami ( Biological Resources and Post-harvest Division )
Yamamizo-Oda Chihiro ( Biological Resources and Post-harvest Division )
Kobayashi Yasufumi ( Biological Resources and Post-harvest Division )
ORCID ID0000-0002-8357-6579KAKEN Researcher No.: 00836968Fujii Kenichiro ( Biological Resources and Post-harvest Division )
ORCID ID0000-0001-7621-5962Nagatoshi Yukari ( Biological Resources and Post-harvest Division )
Fujita Yasunari ( Biological Resources and Post-harvest Division )
Kojiro Tanaka ( Actree Corporation )
Tanaka Tsutomu ( Actree Corporation )
Hiroharu Mizukoshi ( Actree Corporation )
Yasui Yasuo ( Kyoto University )
ORCID ID0000-0003-3804-7886KAKEN Researcher No.: 70293917Yoshikawa Nobuyuki ( Faculty of Agriculture, Iwate University )
KAKEN Researcher No.: 40191556 - ほか
- Publication, etc.
-
Ogata et al. (2021) Frontiers in Plant Science 12: 643499https://doi.org/10.3389/fpls.2021.643499
- Japanese PDF
-
2021_B03_ja.pdf359.53 KB
- English PDF
-
2021_B03_en.pdf382.46 KB
- Poster PDF
-
2021_B03_poster.pdf273.91 KB
* Affiliation at the time of implementation of the study.