Screening of candidate genes for trypanotolerance in mice livers using a DNA microarray
Description
[Objectives]
African trypanosomosis, a protozoan disease transmitted by tsetse flies, is among the most important disease constraints to livestock production, as well as human health, in sub-Saharan Africa. It has been shown that breeds of cattle and inbred strains of mice exhibit genetically based differences in the degree to which they are resistant to the disease. Classical analyses have been made to understand the relation between resistance and host factors specified through characteristic signs and alterations during infection. However, such analyses have failed to elucidate fully the mechanisms of trypanotolerance. In the present study, gene expression levels between mouse strains were comprehensively compared to screen candidate genes for trypanotolerance using a DNA microarray carrying thousands of cDNA fragments.
[Results]
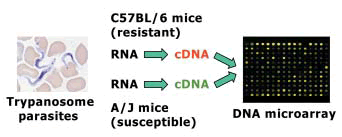
A DNA microarray was prepared by spotting 7,445 species of 65-mer mouse oligonucleotide probes onto a slide glass using an automatic gene arrayer. Resistant C57BL/6 and susceptible A/J mice were infected with trypanosome parasites, and liver samples were obtained on days 0, 4, 7, 10 and 17 after infection. Total RNA was extracted from each tissue, and four pooled RNA samples, each containing five individual samples, were prepared from each mouse strain at each sampling time point. They were then used to synthesize cDNA fragments labeled with red or green fluorescent dye. Red-labeled cDNA of one strain and green-labeled cDNA of the other strain derived from the same sampling time point were mixed and hybridized with the microarray. Following hybridization, signal intensities of both fluorescent dyes bound to each probe were scanned using a laser-confocal array scanner. Differentially expressed genes between C57BL/6 and A/J mice were detected by calculating a ratio of signal intensities from the two mouse strains. Fig. 1 shows a conceptual diagram of DNA microarray analysis.
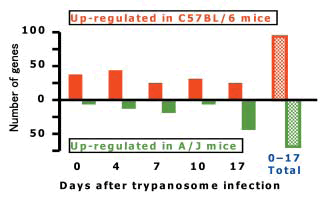
A total of 169 genes were differentially expressed in the livers between the two mouse strains during the course of trypanosome infection (Fig. 2). These included genes encoding important factors involved in a variety of host responses, such as acute phase proteins, complement factors, cytokines and chemokines, electron transporters, intracellular signaling factors, apoptosis factors, ion channels and metabolic enzymes. These genes should be further studied for their roles in conferring genetically based differences in resistance and susceptibility to trypanosome infection among mouse strains.
The present study was conducted under collaboration with the University of Liverpool and the Medical Research Council-UK.
Figure, table
-
Fig. 1. Conceptual diagram of DNA microarray analysis used to screen candidate genes for trypanotolerance. -
Fig. 2. Number of differentially expressed genes in mice liver compared between strains examined using a DNA microarray.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Animal Production and Grassland Division
- Classification
-
Technical A
- Term of research
-
FY2002 (FY2000-2002)
- Responsible researcher
-
NAKAMURA Yoshio ( Animal Production and Grassland Division )
YAGI Yukio ( National Institute of Animal Health )
KITANI Hiroshi ( National Institute of Agrobiological Sciences )
TSUCHIYA Yoshinori ( National Institute of Animal Health )
- ほか
- Publication, etc.
-
Nakamura, Y., Naessens, J., Kierstein, S., Gibson, J. and Iraqi, F. (2003): Comparison of gene expression levels in the liver between resistant and susceptible mouse strains against Trypanosoma congolense infection. Proceedings of the 135th Congress of the Japanese Society of Veterinary Science, Tokyo, Japan, 89.
- Japanese PDF
-
2002_14_A3_ja.pdf774.84 KB
- English PDF
-
2002_14_A4_en.pdf62.76 KB