Monitoring expression profiles of rice genes under cold, drought and high-salinity stresses, and ABA application using both cDNA microarray and RNA gel blot analyses
Description
[Objectives]
Drought, high salinity, and low temperature are the most common environmental stress factors that influence plant growth and productivity. To improve crop yield under conditions of stress, it is important to increase stress tolerance in crops. Genetic engineering can be used as a fast and precise means of achieving enhanced stress tolerance. The identification of novel genes, determination of their expression patterns in response to the stresses, and an improved understanding of their functions in stress adaptation will provide us with the basis for effective engineering strategies to increase stress tolerance. Rice is not only an important crop but also a model monocot crop. Determination of the biological functions of rice genes is one of the greatest challenges of post-genomic research. We have initiated transcriptional monitoring of stress-inducible rice genes in response to dehydration, high salinity, low temperature and abscisic acid (ABA) application.
[Results]
To identify cold, drought, high-salinity and/or ABA-inducible genes in rice, we prepared a rice cDNA microarray including about 1,700 independent cDNAs derived from cDNA libraries prepared from drought-, coldand high-salinity-treated rice plants. We confirmed stress-inducible expression of the candidate genes selected by microarray analysis using RNA gel blot analysis, and finally identified a total of 73 genes as stress-inducible including 57 novel unreported genes in rice. Among them, 36, 62, 57 and 43 genes were induced by cold, drought, high salinity and ABA, respectively (Fig. 1). We observed a strong association in the expression of stress-responsive genes and found 15 genes that responded to all four treatments. Venn diagram analysis revealed greater cross-talk between signaling pathways for drought, ABA and high-salinity stresses than between signaling pathways for cold and ABA stresses, or cold and high-salinity stresses in rice. The rice genome database search enabled us not only to identify possible known cis-acting elements in the promoter regions of several stressinducible genes but also to expect the existence of novel cis-acting elements involved in stress-responsive gene expression in rice stress-inducible promoters. Comparative analysis of Arabidopsis and rice showed that among the 73 stress-inducible rice genes, 62 have already been reported in Arabidopsis with similar functions or gene names, 38 of which have been documented as stress-inducible in Arabidopsis. Transcriptome analysis revealed novel stress-inducible genes suggesting some differences between Arabidopsis and rice in their response to stress.
Figure, table
-
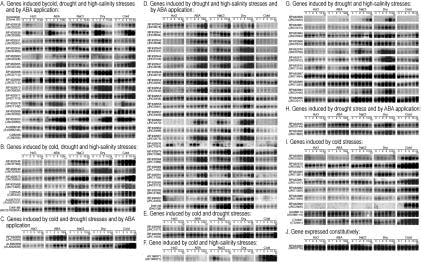
Fig. 1.
RNA gel blot analysis of stress-inducible genes. Each lane was loaded with 10 μg of total RNA isolated from two-week-old rice seedlings that were each exposed to H2O, dehydration, 250 mM NaCl, 100 μM ABA and 4°C cold treatment for 1, 2, 5, 10 and 24 hours. RNA was analyzed by gel-blot hybridization using gene-specific probes based on selected stress-inducible clones obtained via rice cDNA microarray analysis. Stress-inducible clones were classified into various groups on the basis of their expression patterns in RNA gel blot analysis under each stress treatment. Some of the inducible genes were induced by all four stress-treatments while some were upregulated by cold, drought and high salinity, and others were induced by drought and high salinity. Some genes were induced by singular conditions such as cold only.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources Division
- Classification
-
Technical A
- Term of research
-
FY2003 (FY2000-2004)
- Responsible researcher
-
YAMAGUCHI-SHINOZAKI Kazuko ( Biological Resources Division )
ITO Yusuke ( Biological Resources Division )
KATSURA Koji ( Biological Resources Division )
MARUYAMA Kyonoshin ( Biological Resources Division )
RABBANI Malik Ashiq ( Biological Resources Division )
DUBOUZET Joseph Gogo ( Biological Resources Division )
- ほか
- Publication, etc.
-
Dubouzet, J.G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E.G., Miura, S., Seki, M., Shinozaki, K. and Yamaguchi-Shinozaki, K. (2003): OsDREB genes in rice, Oryza sativa L, encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J., 33, 751–763.
Rabbani, M.A., Maruyama, K., Abe, H., Khan, M.A., Katsura, K., Ito, Y., Yoshiwara, K., Seki, M., Shinozaki, K. and Yamaguchi-Shinozaki, K. (2003): Monitoring expression profiles of rice (Oryza sativa L.) genes under cold, drought and high-salinity stresses, and ABA application using both cDNA microarray and RNA gel blot analyses. Plant Physiol., 133, 1755–1767.
Shinozaki, K., Yamaguchi-Shinozaki, K. and Seki, M. (2003): Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol., 6, 410–417.
Yamaguchi-Shinozaki, K. (2002) Japanese Patent Application No. 2002-377316. "Stress-inducing promoter derived from rice".
- Japanese PDF
-
2003_03_A3_ja.pdf1.67 MB
- English PDF
-
2003_03_A4_en.pdf75.21 KB