Yam Variety Identification Toolkit:Step 1
Step 1
In this step, you will determine the most suitable SSR marker sets for your variety identification by:
- Listing all potential Guinea yam varieties/lines in your premises which might mechanically mix with each other.
- Nominating potential SSR marker sets (usually consist of 1 - 3 markers) using the web-application “Minimum SSR Marker Finder for Guinea Yam”.
- Determining the best-bet marker set for analysis.
What is the minimum set of SSR markers?
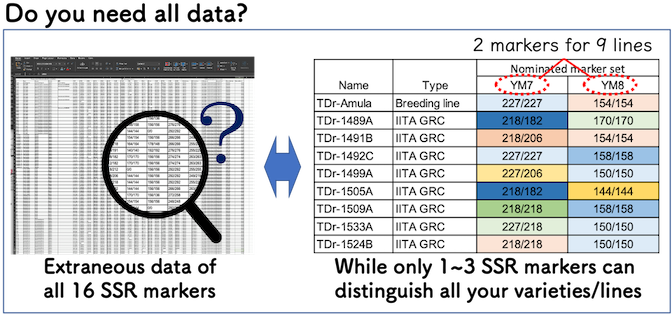
Though currently, 16 SSR markers are available for Guinea yam, you may only need a few markers to distinguish the varieties/lines you have.
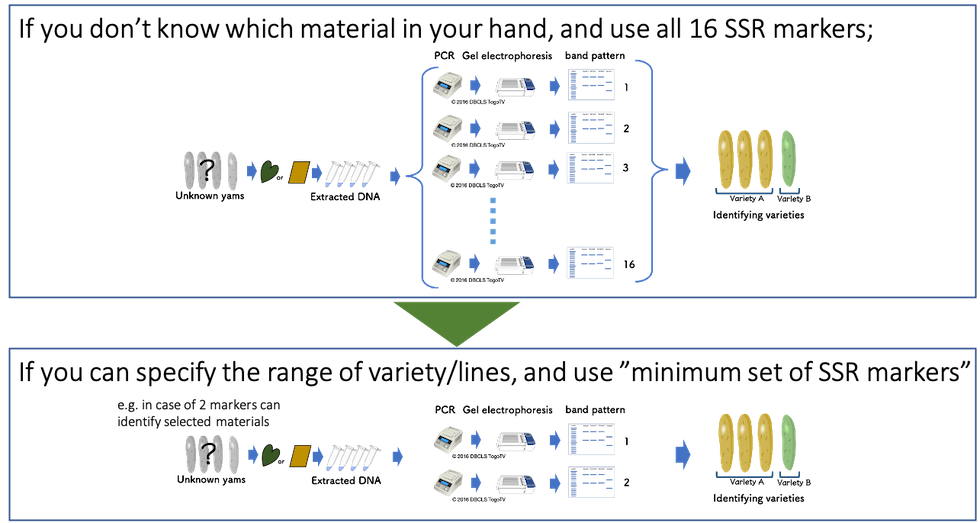
Among the varieties/lines that potentially cause mechanical mixture in your premises/fields, only a few SSR markers (usually 1 - 3 markers) can identify each genotype. The “Minimum SSR Marker Finder for Guinea Yam” is a web-application especially designed to find a minimum set (usually 1 ~ 3 markers) of SSR markers for Guinea yam. This application can assist you to nominate several options of potential SSR marker sets, which can distinguish all varieties/lines you select in a few seconds instead of working on an Excel spreadsheet by your self.
How I can benefit from this web-application?
Using a minimum set of SSR markers (usually 1 - 3 markers), you can potentially, reduce the cost and time up to 1/10 of cases using all 16 SSR markers. Since PCR and gel electrophoresis processes are most cost- and time-consuming steps for variety identification using SSR markers, it drastically saves you time and money.
How to use the web-application
To nominate a minimum SSR marker set using “Minimum SSR Marker Finder for Guinea Yam”, only 4 processes are required:
- Agree with our “Terms of use”
- Nominate the varieties/lines potentially cause mechanical mixture in your premises/fields
- Click “Analysis”
- Select ideal SSR marker set for your variety identification
To get started with the “Minimum SSR Marker Finder for Guinea Yam”, please click here.
For more detailed instruction, please click here.
Limitations
The “Minimum SSR Marker Finder for Guinea Yam” is designed for your variety identification of Guinea yam, but has some limitations:
Case 1: Your materials have to be on the database
The “Minimum SSR Marker Finder for Guinea Yam” requires SSR polymorphism data to be registered in the “Guinea Yam SSR Polymorphism Database”. Currently this database contains the data of approximately 550 genotypes (@Feb 2020) including IITA Genetic Resources Center (GRC) materials, IITA and NARS breeding lines, and local varieties.
To improve this database, we welcome new SSR polymorphism datasets of new genotypes. To support this work, we will be able to assist you for SSR polymorphism analysis in free of charge, till thee closure of EDITS-Yam project in 2021 March. If you have interest, please feel free to communicate with us via our inquiry form.
Case 2: Selection is limited up to 30 varieties/lines
In this web-application, the number of varieties/lines you can select are limited to “up to 30”. This is simply to avoid web-server overload from heavy calculations. If you have more than 30 varieties/lines to be separated, we recommend you consider using the “Minimal Marker” , Perl-based software developed by “Institute of Fruit Tree and Tea Science, NARO (NIFTS, Japan)”.The software can handle large datasets of more than 30 varieties/lines. If you need SSR polymorphism data for this software, please contact us via our inquiry form.
Case 3: No SSR marker set was nominated for your selected varieties/lines
If some of your selected varieties/lines show the exact same polymorphism pattern for all 16 SSR markers available, this “Minimum SSR Marker Finder for Guinea Yam” will not be able to propose a minimum set of SSR markers. This problem may be caused, when:
Case 3-1 "the resolution of 16 SSR marker currently available is not enough to discriminate all materials"
or
Case 3-2 "simply the same variety/line is registered with different names in our database ".
Currently we have no solution for these two possibilities. However, especially in case 3-2, you can use the list of materials that show the same SSR polymorphism pattern to adjust your selection of varieties/lines. If you need our support, please contact us via our inquiry form.
Tips to select the best-bet marker set
The “Minimum SSR Marker Finder for Guinea Yam” can only nominate all suitable SSR marker sets for your selected varieties/lines. Theoretically, any marker sets nominated can be used for your variety identification. However, please understand that you only need one best-bet marker set, though more than a few marker sets can be nominated.
Here are some tips for you to select the best-bet SSR markers. Please check the tips below and select your best marker set among the nominated marker sets. Also, the description of Step 4 will be helpful for your consideration.
- If you have any primers of SSR markers for Guinea yam, select the set that includes the marker(s) you have to reduce the cost.
- Please see "Supplement 1", which indicates the alleles found among 447 Dr accessions by SSR markers for your selection:
- Some markers show large allele size and may require relatively longer loading time for gel electrophoresis.
- Some markers which show larger gaps between allele size may contribute to have better resolution of gel image.
- Some markers with a few alleles may not have wide adaptation for other varieties/lines.
To open suppliment flile iin PDF, please click below link.
- Supplement document
- Supplement 1.pdf68.91 KB
Primer information
Once you choose best-bet marker set, please use below inforrmation for your purchase of primer which will be used in Step 3 "Conduct PCR with nominated markers in step 2, and gel electrophoresis".
| Marker | Primer | Sequence (5' to 3') |
Expected size |
|---|---|---|---|
| YM7 | F | AGCATTGGGTCCTTTCATCC | 203 |
| R | ACAATTCACACAAAGCATGGC | ||
| YM8 | F | TCTTAGGCTTTGGGCAGGG | 166 |
| R | AGTATGCCTACCCTGTTCTTC | ||
|
YM16 |
F | TGAAGAGAATGTTGAGATCGTACC | 150 |
| R | TATCCGGCCCTCTCATTGG | ||
| YM18 | F | GACATTGGGGATCTCTTATCAT | 266 |
| R | TAGCAGCAGTAACGTTAAGGAA | ||
| YM25 | F | GATGGAGATGAGGAGGCCG | 237 |
| R | TTCGAAGCCAGAGCAAGTG | ||
| YM27 | F | TCCAGCTCTTTAGCACAGG | 231 |
| R | AGGAGCATAGGCAACAAGC | ||
| YM28 | F | CCATTCCTATTTAAGTTCCCCT |
333 |
| R | GATGAAGAAGAAGGTGATGATG | ||
| YM30 | F | CCACAACTAAAAACACATGGAC | 212 |
| R | GTGGTAGGGTGTGTAGCTTCTT | ||
| YM31 | F | AAGCCTAGTCGATGGGTGG | 221 |
| R | TGCTGTTCCAACTTCCAAGC | ||
| YM43 | F | GCCTTGTTTTGTTGATGCTTCG | 178 |
| R | CCAGCCCACTAATCCCTCC | ||
| YM44 | F | CGCAACCAGCAAAGGATTTA | 156 |
| R | ATTCTGTCTCTCAAAACCCCT | ||
| YM49 | F | TGGGGTGAGAGAGTAAGTGG | 163 |
| R | TCACCGGGGATCTTCTTGC | ||
| YM50 | F | TTGCCCTTGGGATGTAGGG | 234 |
| R | CATCCCCGTTGTATCCTGC | ||
| YM54 | F | CACTTGCTCTCTCATCGGC | 162 |
| R | TTGACAACCTCTATTTTGCCC | ||
| YM61 | F | AGTGGTGCTGTAGTAACTGGAA | 252 |
| R | CATGACTACCTTTCCTCAATCA | ||
| YM69 | F | CTCTCTACCTCCCAACAAAAAC | 229 |
| R | AATCTTGCACCACCTTTTCTAC |
To open in PDF fle, please click below link.
- Supplement document
- 20210205.pdf40.89 KB