Yam variety identification toolkit
Welcome to our “Yam variety identification toolkit” web site!
The “Yam Variety Identification Toolkit” is an open-access toolkit that enables various users to identify the variety/line of white Guinea yam (Dioscorea rotundata Poir) with simple DNA (SSR) marker techniques.
What’s new!
―――――――
2020.03.16 All function of “Yam variety identification toolkit” is now accessible to all!
―――――――
What is Yam variety identification toolkit?
To meet strong needs of users to identify or confirm the plant materials of guinea yam, the toolkit provides a package of simpler methods to identify the variety/line of white Guinea yam (Dioscorea rotundata Poir) in your laboratory at low cost within a few days. If you want to know more about Guinea Yam, plase visit "About Guinea yam" page.
The expected users of the toolkit are yam researchers, seed growers and inspection officers, and extension officers who are involved in the various steps of yam improvement and dissemination. Please also visit "Use case examle" page to have better image of how each users can be benefitted by our "Yam Variety Identification Toolkit".
Why we need a method to identify variety?
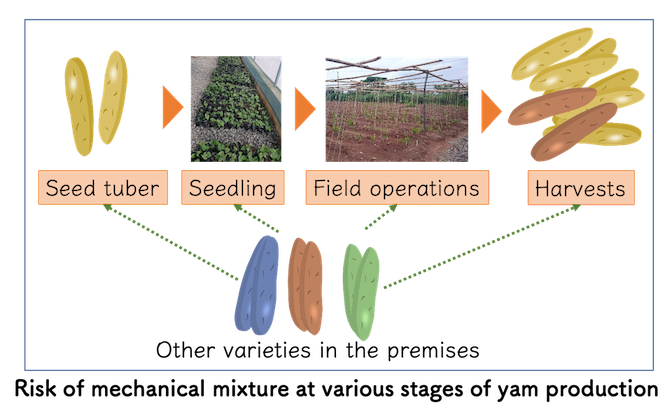
Due to the nature of yam, such as larger seed tuber size, low propagation rate, long growing season and less visible agro-morphological variations, the risk of mechanical mixture and mislabeling of the planting material often burden the works at various stages (planting, vegetative growth, harvesting and storage).
To overcome this risk, the EDITS-Yam project of the Japan International Research Center for Agricultural Sciences (JIRCAS) developed the open-access “Yam Variety Identification Toolkit” for users on our web site.
How to identify variety/lines using SSR markers
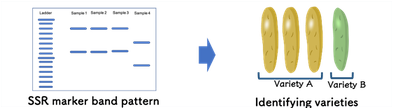
Simple Sequence Repeat (SSR) markers are one of DNA marker systems used for genetic analysis and holds various advantages, such as high reproducibility, low cost and high polymorphism etc. Based on its nature, SSR markers have been used in various work, including genetic profiles identification of the varieties even among very closely related ones.
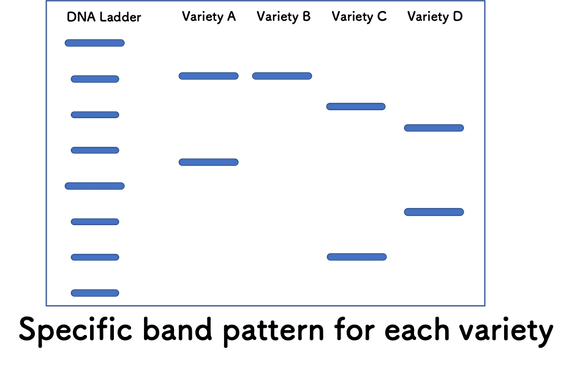
A set of 16 SSR markers is available for Guinea yam (Tamiru et al 2015) and those was used for the genetic diversity analysis (Pachakkil et al 2019) to develop “DrDRS (Dioscorea rotundata Diversity Research Set, 114 lines) for further genetic study selected from available 447 germplasm maintained in IITA. Our earlier work indicates existing 16 SSR markers can differentiate all 114 lines. Using SSR markers and band pattern information (SSR polymorphism pattern) of specific yam variety, you can identify the variety you tested.
How the toolkit supports variety identification of Guinea yam
Does this sound a little complicated? Where can we obtain band pattern information (SSR polymorphism pattern) of my yam variety? Do not worry.
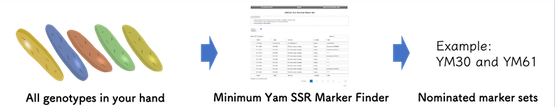
The toolkit supports all steps of yam variety identification with various manuals, videos, and an web-application “Minimum Yam SSR Marker Finder” specially developed for Guinea yam.
* By clicking images, you can jump to each step.
Expected benefits of the Toolkit
The toolkit supports users to verify yam materials in a simple lab facility, at low cost and labor demand, within a few days.
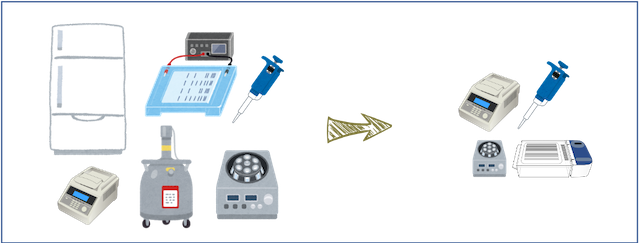
Suitable for small lab facilities
SSR markers do not require a very high-tech facility. However, the methods of the “Yam Variety Identification Toolkit” were reviewed and agreed upon to reduce the requirement of lab facility and large equipment as much as possible, such that a large freezer or liquid N2 for the tissue sample preparation are not required.
Save money
Various tools of “Yam Variety Identification Toolkit” were reviewed and agreed upon to minimize the direct cost (chemicals and consumables) for analysis, as much as possible. Expectedly, the direct cost of analysis can be 1/8 of the conventional methods.
Quicker results
If you are able to conduct the analysis in your lab, the analysis time can be much shorter than shipping out to another analytical lab. Also, the toolkit drastically shortens* the required time of analysis by easy sampling, DNA extraction, and a minimum set of SSR markers compared to the conventional methods.
*when sample number is around 20-30 per analysis.
To maximize the benefits of users
To enable successful variety identification using SSR markers and maximize the benefits of users, below 5 tools are newly developed/proposed under EDITS-Yam project (1st and 2nd phase) of JIRCAS.
- SSR polymorphism database
- Web app “Minimum SSR Marker Finder for Guinea yam”
- DNA extraction method from tuber sample
Also, extensive reviews were conducted for better user experiences, such as:
- Modified C-tab method for DNA extraction from leaf samples
- Leaf and tuber skin sampling method with dried silica gel
- PCR and Gel electrophoresis manuals
- Simple and cost saving Gel electrophoresis equipment
with valuable contributions of our colleagues and partners for the development of this toolkit under our EDITS-Yam of JIRCAS – see “Acknowledgements”