A major and stable quantitative trait locus qSS2 for seed size and shape traits in soybean
Description
Soybean [Glycine max (L.) Merr.] is one of the world’s most economically important food and oil crop. It is a rich source of edible oil and protein, and provides substrate for several important food products. Seed size traits such as length, width, thickness, and single seed weight, and seed shape traits such as length-to-width, length-to-thickness and width-to-thickness, are important determinants of seed yield and appearance quality in soybean. Understanding the genetic architecture of these traits is important to enable their genetic improvement through efficient and targeted molecular breeding by design in soybean, and for the identification of underlying causal genes.To identify quantitative trait locus (QTL) controlling seed size and shape traits in soybean, a recombinant inbred line (RIL) population developed from K099 (small seed size) × Fendou 16 (large seed size) was phenotyped in three growing seasons (2012, 2016, and 2017) in field conditions. A genetic map of the RIL population was developed using 1,485 genotyping by random amplicon sequencing-direct (GRASDi) and 177 SSR markers. QTL analysis was conducted using the QTL IciMapping software. As a result, a total of 53 significant QTLs for seed size traits and 27 significant QTLs for seed shape traits were identified. Six of these QTLs (qSW8.1, qSW16.1, qSLW2.1, qSLT2.1, qSWT1.2, and qSWT4.3) were identified with LOD scores of 3.80–14.0 and R2 of 2.36%–39.49% in at least two growing seasons. Among the above significant QTLs, 24 QTLs were grouped into 11 QTL clusters, with three major QTLs (qSL2.3, qSLW2.1, and qSLT2.1) clustered into a major QTL on chromosome 2, named as qSS2. The effect of qSS2 was validated in a pair of near isogenic lines, and its candidate genes (Glyma.02G269400, Glyma.02G272100, Glyma.02G274900, Glyma.02G277200, and Glyma.02G277600) were mined.
The identified QTLs will pave the way for positional cloning of genes regulating seed size and shape traits and for further understanding of their molecular mechanism in soybean. The results of this study will assist in breeding programs aimed at improving seed size and shape traits in soybean.
Figure, table
-
Fig. 1. Map position of a QTL cluster (qSS2) for seed size and shape traits on chromosome 2 in the K099 × Fendou 16 RIL population
Distances in cM are indicated to the right of the linkage groups and names of markers are shown on the left. qSL2.3, qSLW2.1, and qSLT2.1 represent QTLs for seed length, length-to-width, and length-to-thickness, respectively. -
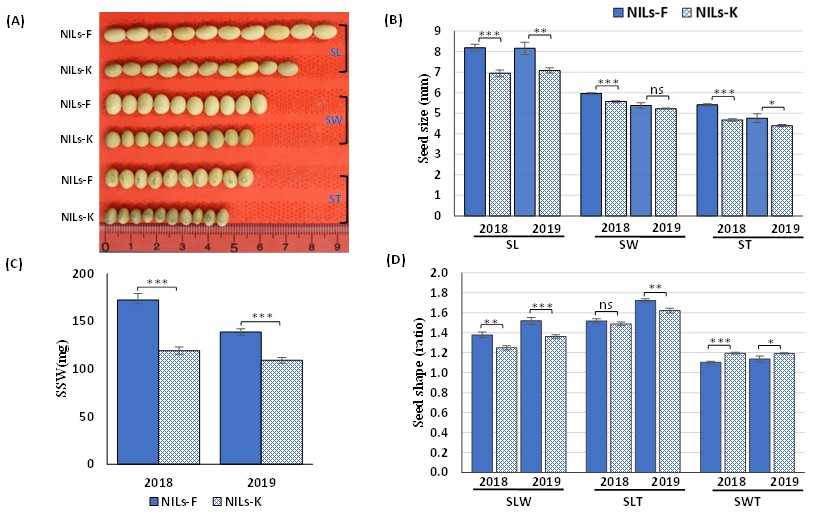
Fig. 2. Seed size and shape phenotypes of near isogenic lines, NILs-F and NILs-K, in 2018 and 2019. (A) phenotypic appearance, (B) SL, seed length; SW, seed width; ST, seed thickness, (C) single seed weight (SSW), and (D) seed shape traits (SLW, ratios of seed length-to-width; SLT, seed length-to-thickness; and SWT, seed width-to-thickness)
Error bars represent means ± SD of three replicates. Asterisks indicate significant differences between NILs-F and NILs-K at 5% (∗), 1% (∗∗), and 0.1% (∗∗∗); ns indicates no significant difference at the 5% level in Student’s t-test.Figures reprinted/modified with permission from Kumawat and Xu (2021)
- Classification
-
Research
- Research project
- Program name
- Term of research
-
FY 2016–2021
- Responsible researcher
-
Xu Donghe ( Biological Resources and Post-harvest Division )
Kumawat Giriraj ( ICAR-Indian Institute of Soybean Research, India )
- ほか
- Publication, etc.
-
Kumawat, G. and Xu, D. (2021) Frontiers in Genetics, 12: 646102https://doi.org/10.3389/fgene.2021.646102
- Japanese PDF
-
2021_B02_ja.pdf419.42 KB
- English PDF
-
2021_B02_en.pdf267.37 KB
- Poster PDF
-
2021_B02_poster.pdf423.9 KB
* Affiliation at the time of implementation of the study.