Identification of quantitative trait locus (QTL) for salt tolerance in soybean
Description
Salt stress, which inhibits soybean germination, plant growth, nodule formation and seed yield, was often reported in soybean-producing areas, especially in the arid and semi-arid areas of developing countries. The agricultural lands are increasingly salinized through saltwater intrusion and use of poor quality irrigation water. Development of soybean varieties with high salt tolerance will provide an efficient way to improve soybean production in a salt stress environment. However, field evaluation and selection for salt tolerance in a soybean breeding program are very difficult because of its complicated genetic nature and the influence of environmental factors. The objectives of this study were: (1) to establish a simple and effective methodology for evaluating a larger number of soybean germplasm for salt tolerance, (2) to perform QTL (quantitative trait locus) analysis for salt tolerant, and (3) to develop DNA markers associated with salt tolerance QTL for marker-assisted selection in a soybean breeding program.
In this study, a simple and effective methodology which enables the evaluation of a larger number of soybean germplasm for salt tolerance was established. Soybean seedlings were each planted in a 17 х 17 cm pot filled with upland field soil. After seven (7) days of emergence, the pots as well as the soybean grown in them were each transformed to large plastic containers (225 х 150 х 45 cm) containing 150 mM NaCl water solution for salt stress by introducing the salt solution into the pots through small holes at the bottom of the pots. The salt solution level in the containers was kept the same as the soil level in the pots to saturate the soil with salt solution, and the salt solution was constantly circulated by an air pump which applies oxygen to the growing soybean. Salt treatment was continued for about four weeks.
Using this evaluation method, more than 600 accessions of wild soybean (Glycine soja) and cultivated soybean (G. max) were evaluated. Several soybean genotypes, such as a Brazilian soybean cultivar, FT-Abyara and a Japanese wild soybean accession, JWS156-1, were identified with high salt tolerance.
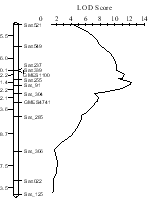
QTL analysis with the mapping populations derived from a cross between FT-Abyara and a salt-sensitive cultivar C01 revealed a major salt tolerant QTL on the soybean linkage group N flanked by SSR markers Sat-91, Satt255, Satt339 and Satt237 (Fig.3). The QTL accounted for 44 % of the total variance for salt tolerance.
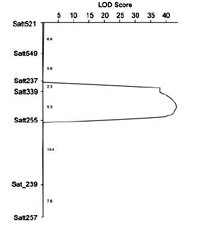
In the mapping populations derived from a cross between the wild soybean accession, JWS156-1 and the salt-sensitive cultivar, Jackson (P1548657), a major QTL which accounted for 68.7% of the total variance for salt tolerance was detected on the same genomic region on linkage group N. Our results indicated that the salt tolerance QTL is conserved in both wild and cultivated soybeans, and the DNA markers associated with the salt tolerance QTL might be used for marker-assisted selection in a soybean breeding program.(Xu, D.H. and Hamwieh, A.)
Figure, table
-
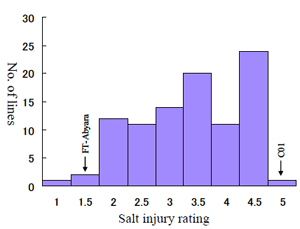
Fig. 1.
Frequency distribution of salt injury rating of the 96 RILs derived from a cross between FT-Abyara (G. max) and a salt -sensitive cultivar C01 (G. max). The salt injury rating values ranged from 1 (normal healthy leaves) to 5 (complete death). -
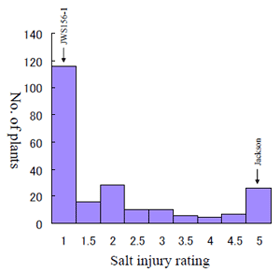
Fig. 2.
Frequency distribution of salt injury rating of the 225 F2 plants derived from a cross between a salt-sensitive soybean cultivar Jackson (G. max) and the wild soybean accession JWS156-1 (G. soja). The salt injury rating values ranged from 1 (normal healthy leaves) to 5 (complete death). -
Fig. 3.
Salt tolerance QTL detected on the linkage group N in the RIL mapping population derived from a cross between FT-Abyara (G. max) and a salt-sensitive cultivar C01 (G. max). -
Fig. 4.
Salt tolerance QTL detected on the linkage group N in the F2 mapping population derived from a cross between a salt-sensitive soybean cultivar Jackson (G. max) and the wild soybean accession JWS156-1 (G. soja).
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources Division
- Classification
-
Technical A
- Term of research
-
FY2008(FY2006~2010)
- Responsible researcher
-
XU Tonghe ( Biological Resources Division )
KAKEN Researcher No.: 90425546HAMWIEH Aladdin ( Biological Resources Division )
- ほか
- Publication, etc.
-
Hamwieh, A. and Xu, D.H. (2008) Conserved salt tolerance quantitative trait locus (QTL) in wild and cultivated soybeans. Breeding Science 58, 355-359.
Hamwieh, A. and Xu, D.H. (2008) QTL analysis for salt tolerance in wild soybean (Glycine soja) . Breeding research 10 Suppl. 1, 156.
Hamwieh, A., Benitez, E.R., Takahashi, R. and Xu, D.H. (2007) Mapping QTL conferring salt tolerance in soybean seedling stage. Breeding research 9 Suppl. 2, 191
- Japanese PDF
-
2008_seikajouhou_A4_ja_Part6.pdf480.56 KB