Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean
Description
Salt-affected soils are generally classified into three main categories depending on the amounts and kinds of salts present. They are saline, sodic (alkaline), and saline-sodic. Both saline and sodic problems are reported to threaten soybean (Glycine max (L.) Merr.) production’s sustainability. Development of soybean cultivars with high salt tolerance will be an effective way to maintain sustainable production of soybean in a salt stress-environment. However, the development of salt-tolerant soybean cultivars in a soybean breeding practice is hampered by the lack of precise evaluation of salt tolerance for breeding lines during the selection process. Therefore, DNA marker-assisted selection (MAS) will be particularly useful in the selection and breeding for salt tolerance. Several studies of quantitative trait locus (QTL) mapping for saline tolerance have been reported, and some DNA markers have been proposed for use in a soybean breeding program to select for salt tolerance. However, little is known about the inheritance of alkaline salt tolerance in soybean. In this study, we identified a QTL allele for alkaline salt tolerance in soybean from a wild soybean accession JWS156-1 (G. soja Sieb. & Zucc.), and developed DNA markers that might be used in a soybean breeding program for alkaline salt tolerance.
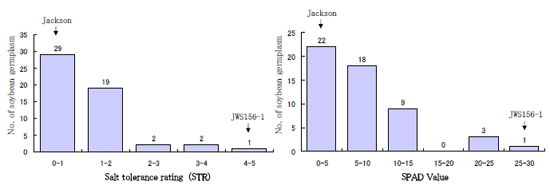
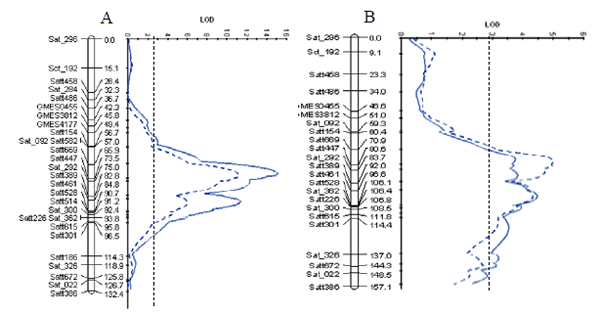
Under alkaline salt treatment with 180 mM NaHCO3 for about three weeks, the wild soybean accession JWS156-1 showed the highest alkaline salt tolerance in terms of salt tolerance rating (STR) and leaf chlorophyll content (SPAD value) among 53 soybean germplasm, including 51 cultivated soybean cultivars and two wild soybean accessions (Fig. 1, 2). To identify QTL for alkaline salt tolerance in soybean, an F6 recombinant inbred line (RIL) mapping population (n = 112) and an F2 population (n = 149) derived from crosses between a cultivated soybean cultivar Jackson and JWS156-1 were used. Evaluation of soybean alkaline salt tolerance was carried out based on salt tolerance rating (STR) and leaf chlorophyll content (SPAD value) after treatment with 180 mM NaHCO3 for about three weeks under greenhouse conditions. In both populations, a significant QTL for alkaline salt tolerance was detected on chromosome 17, which accounted for 50.2 and 13.0% of the total variation for STR in the F6 and the F2 populations, respectively (Fig. 3). The wild soybean contributed the tolerance allele in the progenies. Our results suggest that QTL for alkaline salt tolerance is different from the QTL located on chromosome 3 for NaCl salt tolerance found previously in this wild soybean genotype. The DNA markers closely associated with the QTLs, such as Satt669, Satt447 and Sat_292, might be used for MAS to pyramid tolerance genes in soybean for both saline and alkaline stresses.
Figure, table
-
Fig. 1.
Comparison of alkaline salt tolerance between the wild soybean accession JWS156-1 (right) and the soybean cultivar Jackson (left) after alkaline salt treatment with 180 mM NaHCO3 for about three weeks. -
Fig. 2.
Frequency distribution of salt tolerance rating (STR) and SPAD value of the 53 soybean germplasm. The STR was classified on a scale of one to five, ranging from 1 (complete death) to 5 (normal health leaves). Leaf chlorophyll content (SPAD value) was measured using a chlorophyll meter. -
Fig. 3.
Genetic map of chromosome 17 and QTL LOD score for salt tolerance rating (STR, blue solid line) and leaf chlorophyll content (SPAD, blue dotted line) values in the F6 (A) and the F2 (B) populations derived from the crosses between soybean cultivar Jackson and the wild soybean accession JWS156-1. The vertical dotted lines indicate QTL significant levels (p < 0.01) estimated from a 2000-permutation test by random sampling of phenotypic data.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources Division
- Classification
-
Administration A
- Term of research
-
FY2009~2010
- Responsible researcher
-
XU Tonghe ( Biological Resources Division )
KAKEN Researcher No.: 90425546TUYEN Do Duc ( JIRCAS Visiting Research Fellowship Program )
- ほか
- Publication, etc.
-
https://doi.org/10.1007/s00122-010-1304-y
Tuyen D.D., S.K. Lal, D.H. Xu (2010) Theoretical and Applied Genetics, 121: 229–236.
- Japanese PDF
-
2010_seikajouhou_A4_ja_Part8.pdf88.77 KB