Identification of a quantitative trait locus associated with development of lateral roots in rice employing a genome-wide association study
Description
In recent decades, lowland rice (Oryza sativa L.) cultivation practices have shifted away from the standard system of field puddling and transplanting towards direct seeding. Seedling vigor is an important trait for direct-seeded rice but most modern indica varieties were developed for transplanted rice cultivation and are therefore not expected to have good early seedling vigor. Rapid mobilization of carbon and nutrient reserves stored in the seed is an important determinant of seedling vigor; however, it needs to be supported by the rapid establishment of a root system that can supply additional nutrients and water to the seedling. In rice, nutrient and water uptake is dependent on the abundant presence of lateral roots. Our objective in this study was to map quantitative trait loci (QTL) for early lateral root development and to identify associated candidate genes.
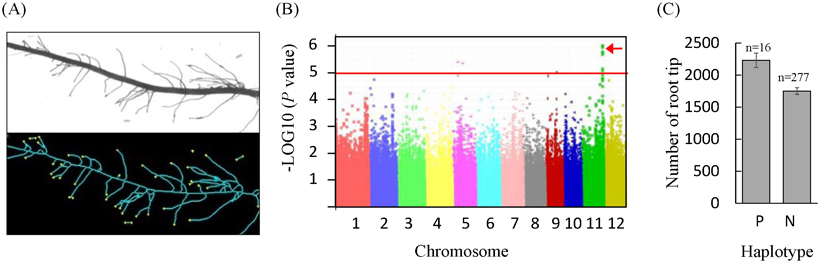
A panel of 307 genotypes including 284 indica genotypes was grown in low-phosphorous nutrient solution for a 14-day period. Digital images of entire root systems were analyzed using the WinRhizo software, which provided an estimate of total root tip number present in the root system (Fig. 1A). As this figure indicates, the vast majority of root tips are tips of fine lateral roots in rice, highlighted here by yellow dots. Using genome-wide association studies (GWAS), we identified one association on chromosome 11 for root tip number (qTIPS-11) (Fig. 1B). The positive haplotype occurred at a low frequency (5.4%) in the panel and increased root tip number by 27.4% (Fig. 1C). The rare nature of the positive haplotype at qTIPS-11 was confirmed in silico within the 3000 sequenced rice genomes (SNP-Seek), of which only 183 accessions, predominantly belonging to the japonica subspecies (71.6%), had the positive haplotype. The positive haplotype was largely absent from indica type accessions.
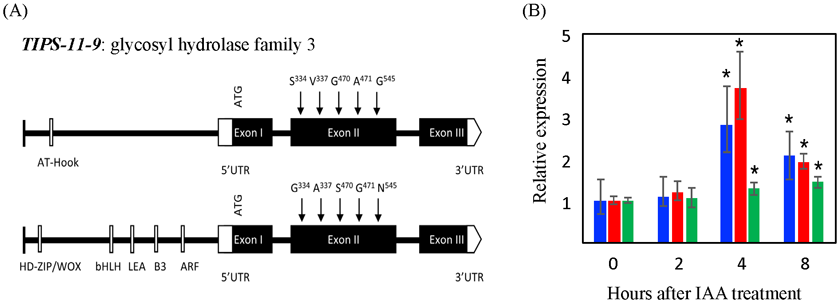
The putative auxin-responsive glucosyl hydrolase (TIPS-11-9; Os11g44950) was identified as a candidate gene (Fig. 2A) because genes responsible for cell wall hydrolysis are needed to facilitate the growth of laterals through several layers of cortical and epidermal cells during root emergence. A T-DNA knock-out mutant of TIPS-11-9 had a 25% reduction in root tip number compared to wild-type, confirming the importance of TIPS-11-9. Two allelic forms of TIPS-11-9 were detected of which the negative allele is missing the auxin response factor (ARF) (Fig. 2A). Expression of TIPS-11-9 was induced by auxin (IAA) addition only in the positive accessions (Fig. 2B), suggesting an allele-specific response of TIPS-11-9 to auxin. Auxin has been implicated in lateral root emergence and development, and thus TIPS-11-9 may be involved in auxin-regulated lateral root formation or development, or both.
Marker-assisted introgression of qTIPS-11 into modern indica varieties will aid in the generation of varieties adapted to direct seeding and to nutrient-limited environments. Clarification of the functions of qTIPS-11 throughout the life cycle of rice will be required.
Figure, table
-
Fig. 1. Detection of a QTL related to lateral root development through GWAS.
(A) Example of digital image analysis of a root segment with scanned image (top) and processed image with root tips indicated by yellow dots (bottom). (B) Manhattan plot indicating significant association between SNP markers on all 12 chromosomes of rice and root tip number. The vertical axis indicates the strength of this association with a threshold set at P = 1.0E-05. (C) Difference between positive (P) and negative (N) haplotypes at qTIPS-11 on root tip number. -
Fig. 2. Characterization of candidate gene TIPS-11-9 for lateral root development at qTIPS-11.
(A) Allelic differences for TIPS-11-9 with negative allele (top) and positive allele (bottom). The positive allele contains an auxin response factor (ARF). (B) Expression of TIPS-11-9 in roots of qTIPS-11 positive accessions Milyang 30 (blue) and Kitaake (red), relative to expression in the negative accession Cauvery (green) in response to auxin (IAA) addition to the rooting medium. Gene expression is shown relative to expression in Cauvery at 0 hours.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Crop, Livestock and Environment Division
Japan International Research Center for Agricultural Sciences Tropical Agriculture Research Front
- Research project
- Program name
- Term of research
-
FY2018(FY2016-FY2018)
- Responsible researcher
-
Wissuwa Matthias ( Crop, Livestock and Environment Division )
KAKEN Researcher No.: 90442722Wang Fanmiao
Ishizaki Takuma ( Tropical Agriculture Research Front )
Tanaka Pariasca-Juan ( Crop, Livestock and Environment Division )
Kretzschmar Tobias ( International Rice Research Institute )
- ほか
- Publication, etc.
-
https://doi.org/10.1111/pce.13400
Wang F et al. (2018) Plant Cell Environ., 41(12):2731-2743
- Japanese PDF
-
A4316.67 KB
A3225.65 KB
- English PDF
-
A4170.45 KB
A3254.14 KB
- Poster PDF
-
2018_B03_poster.pdf282 KB