Complete chloroplast genomes of Erianthus and Miscanthus, and phylogenetic relationships within the Saccharum complex
Description
The genera Erianthus and Miscanthus, both members of the Saccharum complex, are of interest as potential resources for sugarcane improvement and as bioenergy crops. Ongoing studies focus on the conservation and use of wild Erianthus and Miscanthus accessions as breeding materials. However, despite current interest, the taxonomy and phylogenetic relatedness of Saccharum and these related genera have been controversial. Because the chloroplast (cp) genome has conserved gene content and uniparental inheritance, polymorphism within the chloroplast genome is a valuable tool for phylogenetic and evolutionary studies. In this study, we determined the complete cp genome sequence of Erianthus arundinaceus and Miscanthus sinensis. Our analysis of these cp genomes provides insight into the phylogeny and the evolution of the Saccharum complex based on the sequence variations of these cp genomes.
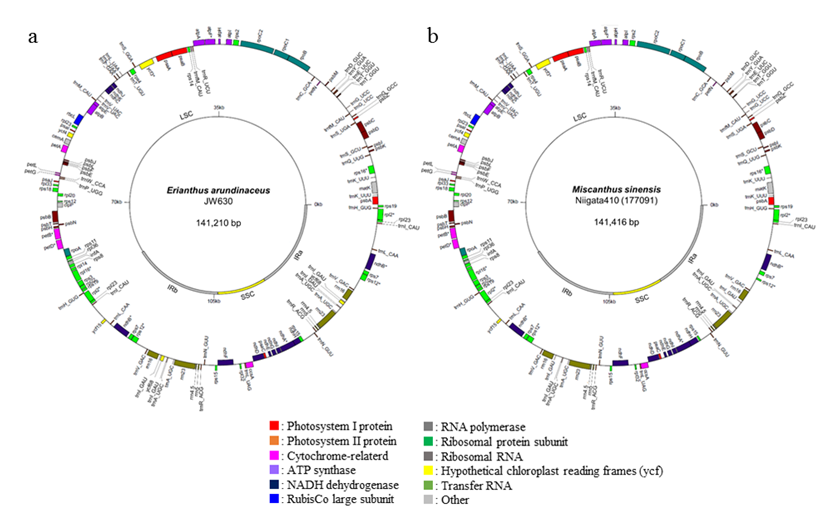
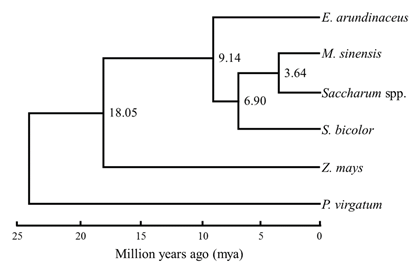
The complete cp genomes of E. arundinaceus (Accession No.: LC160130) and M. sinensis (LC160131) had typical circular structures with 141,210 bp and 141,416 bp in length, respectively (Fig. 1). The number of genes was 143 in E. arundinaceus and 141 in M. sinensis, including 79 and 78 protein-coding genes, respectively. Alignment of the E. arundinaceus and M. sinensis chloroplast genome sequences with the known sequence of S. officinarum demonstrated a high degree of conservation in gene content and order. Using the data sets of 76 chloroplast protein-coding genes, we performed phylogenetic analysis in the Saccharum complex. Our results show that S. officinarum is more closely related to M. sinensis than to E. arundinaceus. We estimated that E. arundinaceus diverged from the subtribe Sorghinae before the divergence of Sorghum bicolor and the common ancestor of S. officinarum and M. sinensis (Fig. 2).
This is the first report of the phylogenetic and evolutionary relationships inferred from maternally inherited variation in the Saccharum complex. Our study provides an important framework for understanding the phylogenetic relatedness of the economically important genera Erianthus, Miscanthus, and Saccharum.
Figure, table
-
Fig. 1. Chloroplast genome maps of Erianthus arundinaceus (a) and Miscanthus sinensis (b).
LSC: large single-copy, SSC: small single-copy, IRa and IRb: inverted repeat a and b. -
Fig. 2. Divergence times of the Saccharum complex estimated based on variation of 76 concatenated protein-coding chloroplast genes
- Affiliation
-
Japan International Research Center for Agricultural Sciences Tropical Agriculture Research Front
- Research project
- Program name
- Term of research
-
FY2017(2011~2020)
- Responsible researcher
-
Tsuruta Shin-ichi ( Tropical Agriculture Research Front )
Ebina Masumi ( Institute of Livestock and Grassland Science, NARO )
Kobayashi Makoto ( Institute of Livestock and Grassland Science, NARO )
MIERUKA ID: 000933Takahashi Wataru ( Institute of Livestock and Grassland Science, NARO )
- ほか
- Publication, etc.
-
https://doi.org/10.1371/journal.pone.0169992
Tsuruta S et al. (2017) PLOS ONE, 12: e0169992
- Japanese PDF
-
A4 259.34 KB
A3 255.98 KB
- English PDF
-
A4 312.85 KB
A3 306.7 KB
- Poster PDF
-
2017_B10_poster.pdf499.4 KB