Evaluations of genetic diversities and population structures of Laotian two small-sized fishes using microsatellite DNA markers
Description
In Indochinese countries, including Laos, greatly diversified indigenous fish fauna is present, and many of them are the important food materials, particularly in the remote rural areas. However, settlements and habitat expansion of invasive alien fishes in recent years and/or agricultural exploitations / urbanization in such areas are now the concerns for decline in species diversity / stock level of indigenous fishes and risk of inbreeding of each species. This situation leads the necessity to investigate the genetic diversities and soundness of each species in the region.
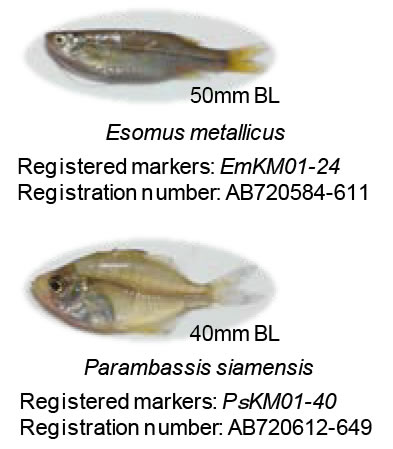
Among the indigenous fish species, Esomus metallicus (Cyprinidae) and Parambassis siamensis (Ambassidae) are the most common and widely distributed species over the Indochina Peninsula (Fig. 1). Both of them are small-sized (60 – 70 mm of maximum standard length) and often utilized in fermented and dried forms. In the present study, we developed the microsatellite DNA markers of these two species collected from 2 sites in Vientiane City and 4 sites in the west coast of Nam Gum River for evaluating the genetic diversities and soundness of local population of fish species.
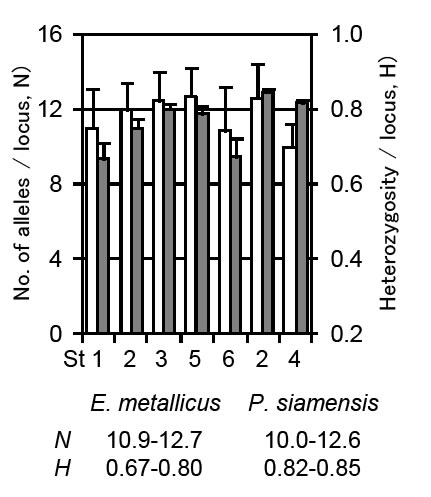
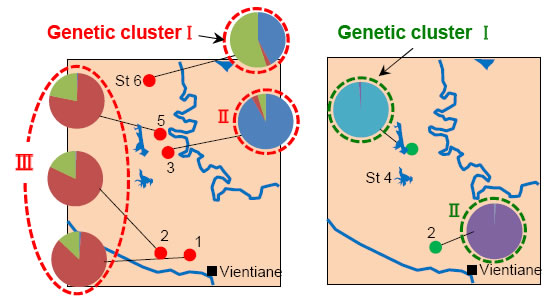
24 and 40 microsatellite DNA markers were developed each for E. metallicus and P. siamensis (Fig. 1). Using these markers, levels of genetic diversities of two species (evaluated based on the number of alleles and heterozygosity) were confirmed to be high enough as of this moment in the investigated area (Fig. 3), and the efficiency of the markers was verified. In addition, based on the genetic cluster analysis of the genotype data for each specimen, the presence of three and two distinct genetic clusters for E. metallicus and P. siamensis was estimated in the investigated area.
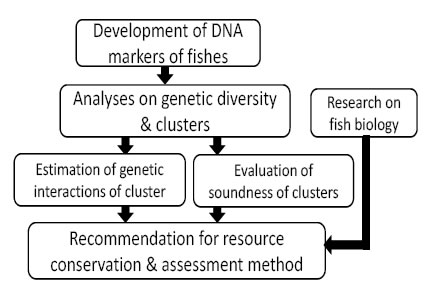
By means of further applications of microsatellite DNA markers over the widespread areas, more detailed genetic characteristics of local populations of the species are confirmable, and the network situation between genetic clusters by migration and breeding can be estimated. Furthermore, this method is applicable not only for the two species above but for the various other species, including the endangered species and the ones for aquaculture for the proper understandings of their strains and management.
Figure, table
-
Fig. 1. Two fish species used in this study and registered DNA markers.
-
Fig. 2. Genetic diversities of two species based on no. of alleles (white bars) and heterozigosity (grey bars).
-
Fig. 3. Structures of genetic clusters of E. metallicus (left) and P. siamenssis (right).
-
Fig. 4. Significance in usage of DNA markers for resource conservation & sustainable exploitation.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Fisheries Division
- Classification
-
Administration B
- Research project
- Program name
- Term of research
-
FY 2013 (FY 2011-FY 2015)
- Responsible researcher
-
Morioka Shinsuke ( Fisheries Division )
KAKEN Researcher No.: 40455259Koizumi Noriyuki ( Institute for Rural Engineering, NARO )
KAKEN Researcher No.: 60301222Vongvichith Bounsong ( Living Aquatic Resources Research Center, Lao PDR )
- ほか
- Publication, etc.
-
Koizumi, N. et al. (2012) Conservation Genetics Resources, 4: 1027-1030.
Koizumi, N. et al. (2012) Conservation Genetics Resources, 4: 1031-1035.
Koizumi, N. et al. (2012) PAWEES 2012.
- Japanese PDF
-
2013_C01_A3_ja.pdf223.16 KB
2013_C01_A4_ja.pdf381.34 KB
- English PDF
-
2013_C01_A3_en.pdf166.79 KB
2013_C01_A4_en.pdf790.66 KB
- Poster PDF
-
2013_C01_poster.pdf365.37 KB