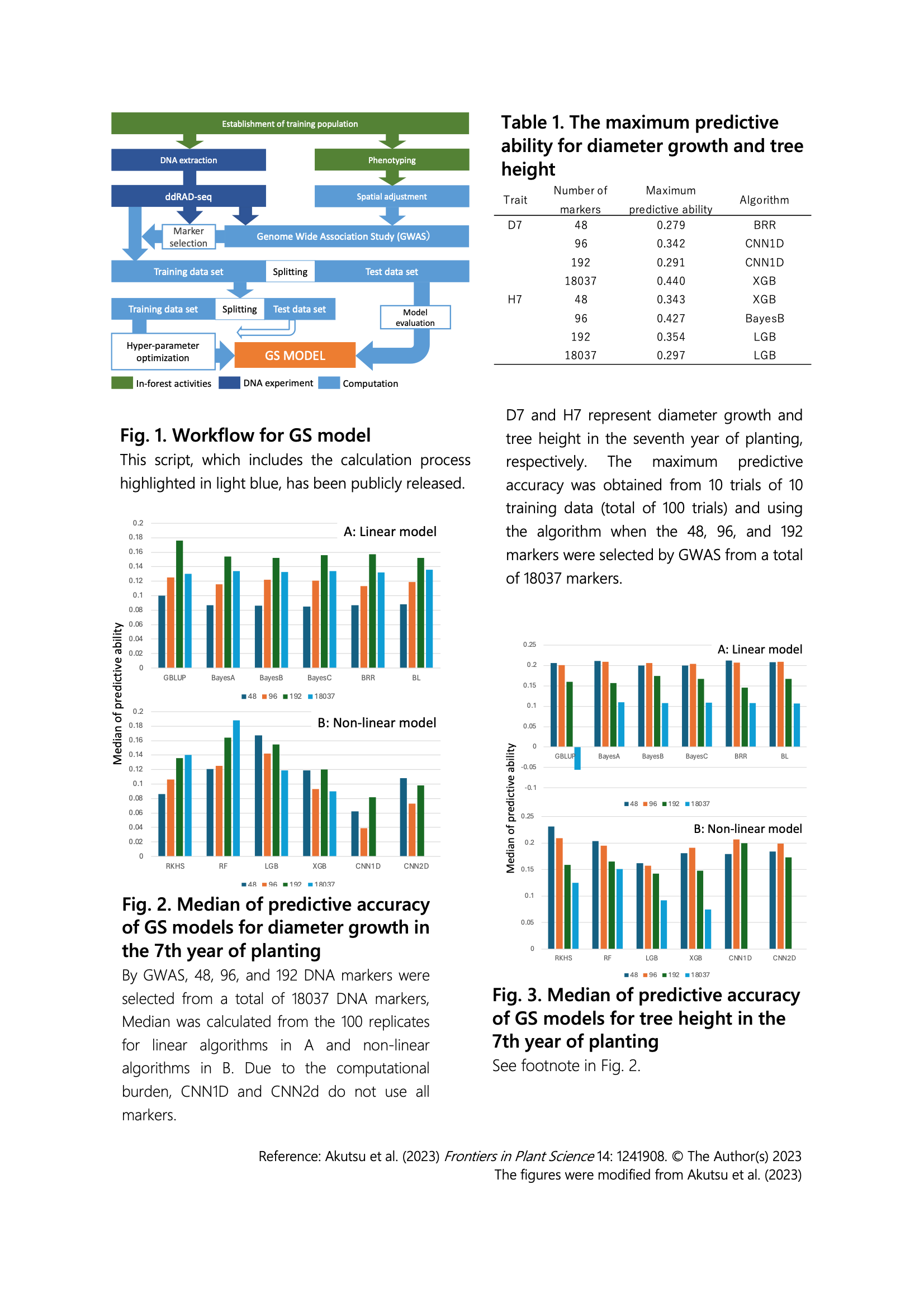
Development of models to predict stem diameter and tree height of Shorea platyclados (Dipterocarpaceae) based on genomic information from seedlings
Description
In recent years, rapid advancements in DNA sequencing technology have made it relatively inexpensive to collect genomic information even for woody plants that are close to wild species. However, for woody plants with large individual sizes and long lifespans, evaluating the performance of next generations (known as progeny trial) after breeding superior individuals requires vast land and time for their growth. This becomes a limiting factor in selecting superior individuals. Therefore, attention is now focused on methods that utilize genomic information to predict specific phenotypes. Specifically, if we can predict future phenotypes using genomic information from offspring, it becomes possible to overcome previous limiting factors. In this study, we aimed to develop a genomic selection model for Shorea macrophylla, a tree species in the Dipterocarpaceae, which is expected to be intensively planted in Southeast Asian tropical forests.We established a workflow using a training population of a progeny trial forest with relatively less environmental heterogeneity for genomic selection (GS) model. We collected genomic information and focal phenotypes. Using these data, we corrected the spatial structure of the phenotypes due to environmental heterogeneity, detected important genetic markers by genome-wide association study (GWAS), and established a workflow for developing GS models (Fig. 1). The workflow was implemented using scripts in R and Python, which we have made publicly available. By selecting highly correlated markers based on GWAS results, we achieved usable predictive accuracy using both linear models (6 algorithms) and nonlinear models (6 algorithms). With this model, it becomes possible to select offspring based on genomic information from the training population without the subsequent progeny testing (Table 1). This approach promises to enhance breeding outcomes related to growth. Specifically, when predicting the 7-year diameter growth, we calculated the median of predictive accuracy for 100 randomly generated models using linear and nonlinear methods. Given that diameter growth results from a combination of primary and secondary growth, the use of nonlinear models, which can account for interactions among numerous genes, yielded higher predictive accuracy (Fig. 2). Conversely, tree height, being only influenced by primary growth, was associated with fewer relevant genes. Therefore, narrowing down the number of associated genes through GWAS led to a more accurate GS model (Fig. 3).
Figure, table
- Research project
- Program name
- Term of research
-
FY2021-2023
- Responsible researcher
-
Tani Naoki ( Forestry Division )
KAKEN Researcher No.: 90343798Na'iem Mohammad ( Gadjah Mada University )
Widiyatno ( Gadjah Mada University )
Indrioko Sapto ( Gadjah Mada University )
Sawitri ( Gadjah Mada University )
AKUTSU Haruto ( University of Tsukuba )
Tsumura Yoshihiko ( University of Tsukuba )
KAKEN Researcher No.: 20353774 - ほか
- Publication, etc.
-
Akutsu et al. (2023) Frontiers in Plant Science, 14:1241908.https://doi.org/10.3389/fpls.2023.1241908
- Japanese PDF
-
2023_A05_ja.pdf514.72 KB
- English PDF
-
2023_A05_en.pdf265.49 KB
* Affiliation at the time of implementation of the study.