Clarifying the genetic diversity of Amaranthus tricolor ‘Hiyuna,’ a traditional Asian vegetable
Description
Amaranthus tricolor L., called ‘hiyuna’ in Japanese, is used as a traditional leafy vegetable in Asia. A. tricolor is resistant to major diseases, is more tolerant to environmental stresses, and is a rich source of nutrients such as iron and calcium, as well as vitamin C and beta-carotene. Yet despite this diversity and excellent nutritional quality, breeding of improved cultivars is widely neglected in modern breeding. A. tricolor accessions are conserved in the World Vegetable Center (WorldVeg) genebank (https://avrdc.org) and in the USDA National Plant Germplasm Information System (https://www.ars-grin.gov). These accessions hold a wide variety of genetic variations and useful agronomic traits with a high potential for breeding improved cultivars (Fig. 1). Some studies have reported on the evaluation of A. tricolor genetic resources, but the diversity conserved in both genebanks has not yet been systematically evaluated.
In this study, we evaluated the genetic diversity in the collection of A. tricolor accessions conserved by the WorldVeg and USDA genebanks based on genome-wide single-nucleotide polymorphisms (SNPs) developed using double-digest RAD-Seq (ddRAD-Seq), and created a core collection, which is valuable in improving crop breeding programs for performing extensive evaluations with minimal materials.
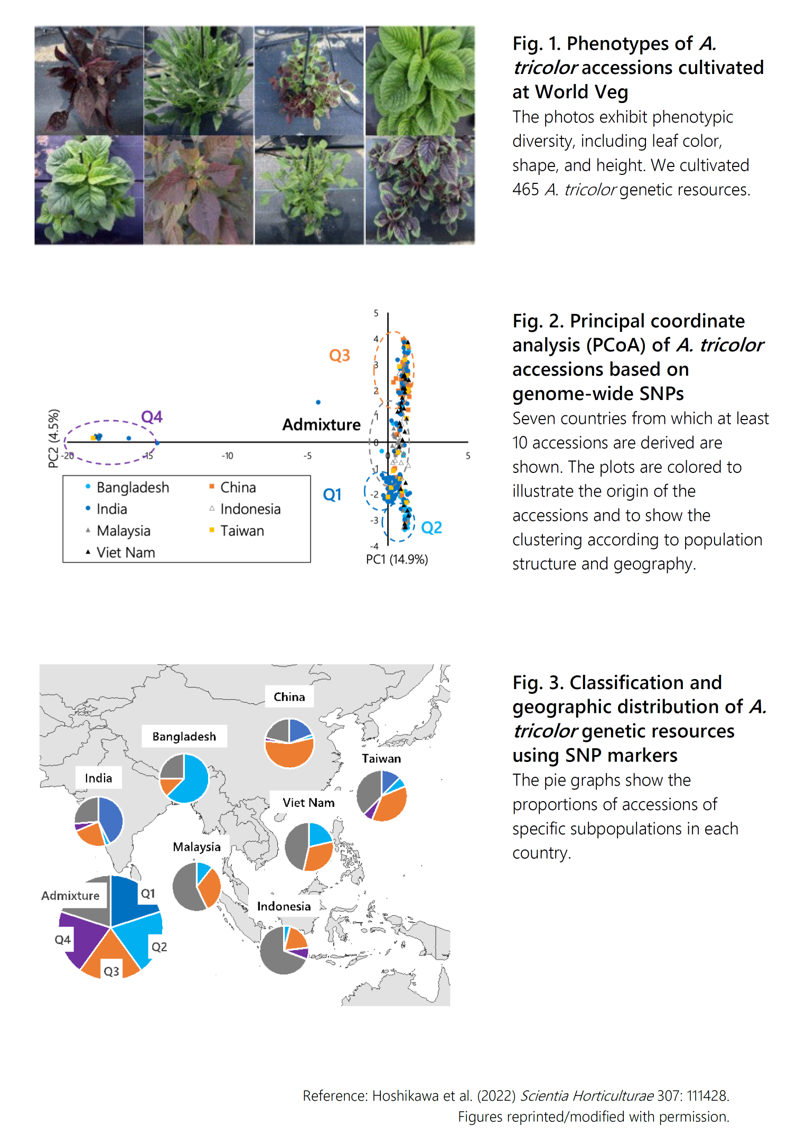
We analyzed the genetic diversity and population structure among 465 A. tricolor accessions with SNPs developed by ddRAD-Seq. We identified a set of 5,638 SNPs without missing data in 440 accessions in order to establish a breeding platform. We analyzed genetic diversity by principal coordinate analysis (PCoA) of the accessions using this marker set. The 377 A. tricolor accessions clustered into 4 main subpopulations (Q1–Q4) and an admixture group (Fig. 2). The proportion of accessions from India, Bangladesh, and China in Q1, Q2, and Q3 was significantly higher than that in other countries, and the proportion of the admixture group in all accessions was highest in accessions from Southeast Asia, especially Indonesia and Malaysia (Fig. 3). In addition, we created a core collection of 105 accessions representing the genetic diversity of 377 source accessions. This core collection is available for research and breeding through the WorldVeg genebank.
Marker selection breeding using the SNP markers and core collection obtained this time will pave the way for the development of breeding techniques and new varieties to improve nutritional value, eating quality, and yield. It is also expected to contribute to sustainable vegetable production in tropical and subtropical regions.
Figure, table
- Research project
- Program name
- KAKEN
- Term of research
-
FY2019–2022
- Responsible researcher
-
Hoshikawa Ken ( Biological Resources and Post-harvest Division )
KAKEN Researcher No.: 70634715Yoshioka Yosuke ( University of Tsukuba )
Shirasawa Kenta ( Kazusa DNA Research Institute )
ORCID ID0000-0001-7880-6221KAKEN Researcher No.: 60527026 - ほか
- Publication, etc.
-
Hoshikawa et al. (2022) Scientia Horticulturae 307: 111428https://doi.org/10.1016/j.scienta.2022.111428
- Japanese PDF
-
2022_B07_ja.pdf881.65 KB
- English PDF
-
2022_B07_en.pdf447.94 KB
* Affiliation at the time of implementation of the study.