Whole genome sequence of Guinea yam (Dioscorea rotundata ) and development of a DNA marker for sex determination
Description
Yam (Dioscorea spp.), a widely cultivated tuber crop in Africa, Asia, and South America, serves as a key food source for people in those regions. However, yam breeding has remained ineffective, constrained by the crop’s unique inherent attributes including a long growth cycle, dioecy (i.e., having separate male and female plants among varieties), polyploidy, high heterozygosity, and inconsistent year-to-year fluctuations of various traits. Regarding crop characteristics, the use of advanced genetic tools and resources retains enormous potential to boost the breeding efficiency of yam.
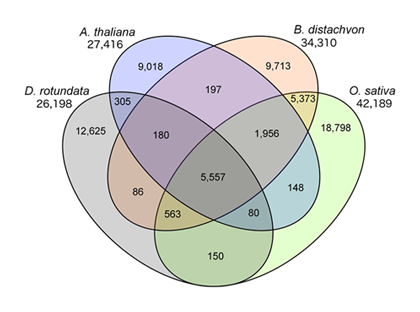
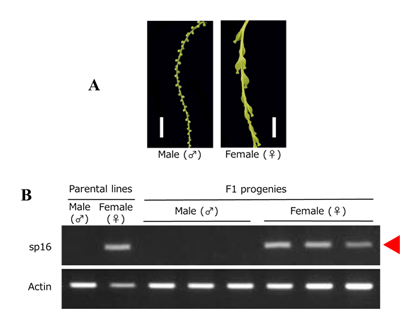
As an initial attempt, we focused on Guinea yam (D. rotundata), which is one of the most important species for regional food security and farmer income generation especially in West Africa (Fig. 1). Based on our genome analysis of D. rotundata, a 594 Mb of genome sequence was successfully assembled. The results of gene prediction using the genome sequence showed a total of 26,198 genes in D. rotundata (Fig. 2). By comparison with other known model plant species, 5,557 D. rotundata genes were identified with orthologous relationship to the B. distachyon, O. sativa, and A. thaliana gene models. On the other hand, 12,625 D. rotundata genes were uniquely found in D. rotundata (Fig. 2). The Dioscorea genus is characterized by the occurrence of separate male and female plants (dioecy), a feature that has limited efficient yam breeding. To infer the genetics of sex determination, we performed whole-genome resequencing of bulked segregants (quantitative trait locus sequencing [QTL-seq]) in F1 progeny segregating for male and female plants, and identified a genomic region associated with sex determination. Based on the result, a molecular marker for sex identification of D. rotundata was developed, enabling us to discriminate male and female plants at the seedling stage (Fig. 3).
The genome sequence information obtained in this work is expected to greatly accelerate the genetic analysis of D. rotundata and relative yam species. While the use of a DNA marker for sex identification should immediately contribute to yam breeding for selection of suitable parental materials at the seedling stage, the effectiveness of the QTL-seq approach demonstrated in this study in outcrossing crops and organisms with highly heterozygous genomes should likewise boost the application of genetic analysis in orphan crops such as yam. This in turn could help contribute to food security and improve the sustainability of tropical agriculture.
Figure, table
-
Fig. 1. Yam cultivation field (left) and tubers being sold in the market (right).
-
Fig. 2. Venn diagram showing conserved and unique genes at 1:1 correspondence among D. rotundata, Arabidopsis thaliana, Brachypodium distachyon, and Oryza sativa. Total gene counts in each genome are given below the species name.
-
Fig. 3. Sex discrimination by the DNA marker developed in this study. A: Male and female inflorescence of D. rotundata. Bars = 10 mm. B: Results of agarose gel electrophoresis of PCR products amplified by DNA marker sp16 (sp16). Actin from D. rotundata (Dr-Actin) served as a control to show that template DNA was present for all samples.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Tropical Agriculture Research Front
- Research project
- Program name
- Term of research
-
FY2017(2011~2015)
- Responsible researcher
-
Yamanaka Shinsuke ( Tropical Agriculture Research Front )
MIERUKA ID: 001785Muranaka Satoru ( Research Strategy Office )
Takagi Hiroko ( Tropical Agriculture Research Front )
Tamiru Muluneh ( Iwate Biotechnology Research Center )
Terauchi Ryohei ( Iwate Biotechnology Research Center )
Asideu R ( International Institute of Tropical Agriculture )
- ほか
- Publication, etc.
-
https://doi.org/10.1186/s12915-017-0419-x
Tamiru M et al. (2017) BMC Biology, 15:86
- Japanese PDF
-
A4 348.89 KB
A3 398.95 KB
- English PDF
-
A4 235.21 KB
A3 161.81 KB
- Poster PDF
-
2017_B02_poster.pdf403.55 KB