Draft genome sequence of an inbred line of Chenopodium quinoa, an allotetraploid pseudocereal crop with high nutritional properties and tolerance to abiotic stresses
Description
Chenopodium quinoa (quinoa) is an annual herbaceous plant that originated from the Andes region of South America. It is a pseudocereal crop of the Amaranthaceae family, which also includes spinach (Spinacia oleracea) and sugar beet (Beta vulgaris). Quinoa is emerging as a key crop with the potential to contribute to global food security, and is considered to be an optimal food source for astronauts due to its great nutritional profile and ability to tolerate adverse environments such as high salinity. In addition, plant virologists utilize quinoa as a representative diagnostic host to identify virus species.
The major cultivation area of quinoa ranges from Columbia to central Chile, and includes altitudes from 0 m up to 4,000 m above sea level receiving an annual amount of rainfall of 80mm to 2,000mm. Quinoa exhibits great tolerance to soil salinity, frost, and drought, thus it is well suited for growing under unfavorable climatic and environmental conditions and. Moreover, quinoa is an excellent nutritional source of various minerals (e.g., Ca, Fe, P, and Zn), vitamins (e.g., A, B1, B2, C, and E), linolenate, natural antioxidants such as polyphenols, dietary fiber, and high-quality protein containing high levels of essential amino acids. Being gluten-free, quinoa is suitable for consumption by individuals who are allergic or intolerant to wheat, rye, and barley. Because of the great nutritional value of quinoa seeds and the high adaptability of quinoa plants to hostile environments, quinoa is deemed by the Food and Agriculture Organization of the United Nations (FAO) to be an important crop with the potential to contribute to food security worldwide. Moreover, the USA’s National Aeronautics and Space Administration (NASA) considers quinoa as an optimal food source for astronauts on long-term space missions in isolated conditions. However, molecular analysis of quinoa is restricted by its genome complexity derived from allotetraploidy and its genetic heterogeneity due to outcrossing.
To overcome these limitations, we established the inbred and standard quinoa accession Kd that allows molecular analysis to unravel the mechanism of its high nutritional value, tolerance to unfavourable environments, and susceptibility to a broad range of viruses, and provided the draft genome sequence of Kd using an optimized combination of high-throughput next generation sequencing on the PacBio RS II and Illumina Hiseq 2500 sequencers. The de novo genome assembly contained 25 k scaffolds consisting of 1 Gbp with N50 length of 86 kbp. Based on these data, we constructed the free-access Quinoa Genome DataBase (QGDB; http://quinoa.kazusa.or.jp), which provides annotations of in silico predicted genes. Furthermore, we utilized comparative genomics and experimental approaches to identify genes in quinoa that are involved in abiotic and biotic stress responses. Thus, these findings yield insights into the effect of allotetraploidy on genome evolution and the mechanisms underlying agronomically important traits of quinoa.
Figure, table
-
Fig. 1. Morphological characteristics of quinoa (Kd) plants.
(A) Dried mature quinoa (Kd) seeds. (B, C, D) 6-, 8-, and 16-week-old quinoa (Kd) plants grown in soil. (E) Head of 17-week-old quinoa plants at harvest time. -
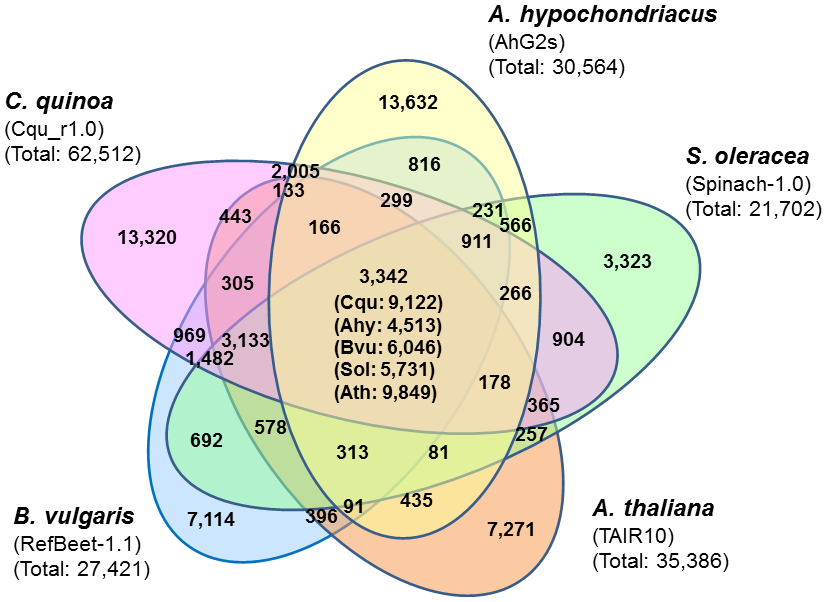
Fig. 2. Cluster analysis of the 62,512 filtered gene sequences.
Predicted genes in Chenopodeum quinoa, Amaranthus hypochondriacus, Beta vulgaris, Spinacia oleracea, and Arabidopsis thaliana were clustered into gene families. In this analysis, we used the filtered dataset of quinoa consisting of 62,512 sequences annotated by performing BLASTP searches against the NCBI’s NR database. The number in each section represents the number of clusters, and the numbers in parentheses in the center section represent the numbers of genes included in the analysis from each species. The number below the species name marks the total number of genes used as input for CD-hit (-c 0.4, -aL: 0.4).
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources and Post-harvest Division
- Research project
- Program name
- Term of research
-
FY2016(FY2015~FY2016)
- Responsible researcher
-
Fujita Yasunari ( Biological Resources and Post-harvest Division )
Yasui Yasuo ( Kyoto University )
ORCID ID0000-0003-3804-7886KAKEN Researcher No.: 70293917Hirakawa Hideki ( Kazusa DNA Research Institute )
ORCID ID0000-0002-7128-7390KAKEN Researcher No.: 80372746Mori Masashi ( Ishikawa Prefectural University )
KAKEN Researcher No.: 00320911Tanaka Tsutomu ( Actree Corporation )
- ほか
- Publication, etc.
-
https://doi.org/10.1093/dnares/dsw037
Yasui Y et al. (2016) DNA Research, 23(6): 535-546 [Editor’s Choice]
Quinoa Genome DataBase (QGDB): http://quinoa.kazusa.or.jp
- Japanese PDF
-
A4 395.32 KB
A3 293.93 KB
- English PDF
-
A4 842.04 KB
A3 791.95 KB
- Poster PDF
-
Poster 482.96 KB