Integrated analysis of the effects of dehydration and cold on rice metabolites, phytohormones, and gene transcripts
Description
Land plants must mount suitable responses to overcome the adverse effects of water stress caused by either drought or low-temperature conditions. Among external stresses, water stress is one of the most important limitations to crop productivity. Discoveries of useful genes for molecular breeding using metabolomics and transcriptomics promise to facilitate the improvement of crop yields under water stress conditions. It is important to identify plant metabolites and transcripts that respond to water stress to determine the essential steps in molecular processes related to the effective adaptation of plants to stress conditions. Metabolomic and transcriptomic data have provided much information on the metabolite, phytohormone, and transcript networks that control plant growth and development. Rice is important not only as a major crop but also as a model monocot.
In this study, we performed an integrated analysis of the metabolites, phytohormones, and gene transcripts in rice plants subjected to dehydration or cold treatments. Our aim was to comprehensively survey the molecular responses of rice to dehydration or cold stimuli. We used three types of MS systems: gas chromatography coupled with time-of-flight MS (GC-TOF-MS), capillary electrophoresis coupled with MS (CE-MS), and liquid chromatography coupled with MS (LC-MS). We identified and characterized representative dehydration-responsive and cold-responsive metabolites and phytohormones. We also performed a transcriptome analysis using a rice oligonucleotide microarray. We analyzed metabolite–gene and phytohormone–gene correlations and identified several genes encoding metabolic enzymes that might play key roles in the responses of rice plants to dehydration or cold. We compared the roles of the identified rice genes with those of their counterparts in Arabidopsis under dehydration or cold conditions.
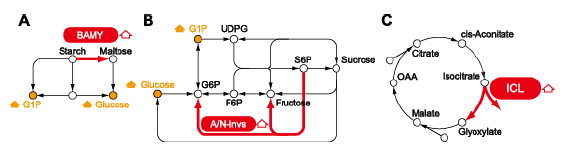
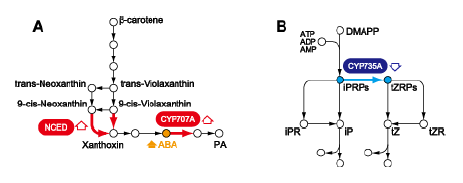
An integrated analysis of metabolites and gene expression indicated that several genes encoding enzymes involved in starch degradation, sucrose metabolism, and the glyoxylate cycle are upregulated in rice plants exposed to dehydration or cold, and that these changes are correlated with the accumulation of glucose, fructose, and sucrose (Fig.1). In particular, high expression levels of genes encoding isocitrate lyase and malate synthase in the glyoxylate cycle correlate with increased glucose levels in rice, but not in Arabidopsis, under dehydration conditions, indicating that the regulation of the glyoxylate cycle may be involved in glucose accumulation under dehydration conditions in rice, but not in Arabidopsis. An integrated analysis of phytohormones and gene transcripts revealed an inverse relationship between abscisic acid (ABA)-signaling and cytokinin-signaling under cold and dehydration stresses; these stresses increase ABA signaling and decrease cytokinin signaling (Fig. 2). High levels of OsNCED transcripts correlate with ABA accumulation, and low levels of CYP735A transcripts correlate with decreased levels of a cytokinin precursor in rice. This reduced expression of CYP735As occurs in rice, but not in Arabidopsis. Therefore, transcriptional regulation of CYP735As might be involved in regulating cytokinin levels under dehydration and cold conditions in rice, but not in Arabidopsis.
Figure, table
-
Fig. 1. Carbohydrate and amino acid metabolic pathways. (A) Starch degradation; (B) Sucrose metabolism; (C) Glyoxylate cycle: BAMY, β-amylase; A/N-invs, alkaline/neutral invertase; ICL, isocitrate lyase.
-
Fig. 2. Pathways for ABA and CK biosyntheses. (A) ABA biosynthesis; (B) CK biosynthesis. NCED, 9-cis-epoxycarotenoid dioxygenase.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources and Post-harvest Division
- Classification
-
Administration A
- Research project
- Program name
- Term of research
-
FY 2014 (FY 2010-FY 2014)
- Responsible researcher
-
Maruyama Kyounoshin ( Biological Resources and Post-harvest Division )
Shinozaki Kazuko ( University of Tokyo )
ORCID ID0000-0002-0249-8258 - ほか
- Publication, etc.
-
https://doi.org/10.1104/pp.113.231720
Maruyama et al. (2014) Plant Physiol. 164(4): 1759-1771.
- Japanese PDF
-
2014_B03_A3_ja.pdf218.35 KB
2014_B03_A4_ja.pdf202.01 KB
- English PDF
-
2014_B03_A3_en.pdf50.57 KB
2014_B03_A4_en.pdf57.35 KB
- Poster PDF
-
2014_B03_poster.pdf274.39 KB