Metabolic networks involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A
Description
Low temperature and dehydration are adverse environmental conditions that affect plant growth and productivity. Many genes have been described that respond to both stresses at the transcriptional level, and their gene products are thought to function in stress tolerance and response even though these stresses are quite different. These genes include key metabolic enzymes, late embryogenesis-abundant (LEA) proteins, detoxification enzymes, chaperones, protein kinases, and transcription factors.
Overexpression of DREB1/CBFs driven by the CaMV 35S promoter increases stress tolerance to freezing, dehydration, and high salinity in transgenic Arabidopsis. More than 40 downstream targets of DREB1A/CBF3 have been identified by microarrays. Overexpression of the constitutively active form of DREB2A (35S:DREB2A-CA) significantly increases dehydration tolerance but only slightly increases freezing tolerance. Microarray analyses of the 35S:DREB2A-CA plants revealed that DREB2A regulates expression of many dehydration-responsive genes. However, some genes regulated by DREB1A are not regulated by DREB2A.
DREB1A/CBF3 and DREB2A are transcription factors that specifically interact with a cis-acting dehydration-responsive element (DRE), which is involved in cold- and dehydration-responsive gene expression in Arabidopsis. Overexpression of DREB1A improves stress tolerance to both freezing and dehydration in transgenic plants. In contrast, overexpression of an active form of DREB2A results in significant stress tolerance to dehydration, but only slight tolerance to freezing in transgenic plants. The downstream gene products for DREB1A and DREB2A are reported to have similar putative functions, but downstream genes encoding enzymes for carbohydrate metabolism are very different between DREB1A and DREB2A. We demonstrate that under cold and dehydration conditions, expression of many genes encoding starch-degrading enzymes, sucrose metabolism enzymes, and sugar alcohol synthases changes dynamically, and consequently many kinds of monosaccharides, disaccharides, trisaccharides and sugar alcohols accumulate in Arabidopsis. We also show that DREB1A overexpression can cause almost the same changes in these metabolic processes and that these changes seem to improve freezing and dehydration stress tolerance in transgenic plants. In contrast, DREB2A overexpression did not increase the level of any of these metabolites in transgenic plants. Strong freezing stress tolerance of the transgenic plants overexpressing DREB1A may depend on accumulation of these metabolites.
Figure, table
-
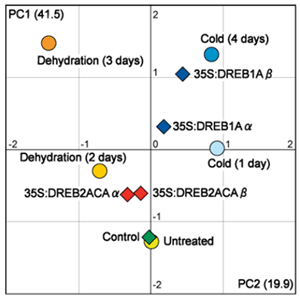
Fig.1.
Statistical analyses of metabolite profiles: We analyzed two independent lines of 35S:DREB1A (a and b) and 35S:DREB2A-CA (a and b) plants. The levels of metabolites for both DREB1A and DREB2A in each b line were higher than those in each a line. Principal component analysis of metabolites: The y- and x-axes are PC1 and PC2, respectively. The solid circles indicate untreated, cold-exposed, and dehydration-exposed plants. The solid diamonds represent control, 35S:DREB1A and 35S:DREB2A-CA plants. -
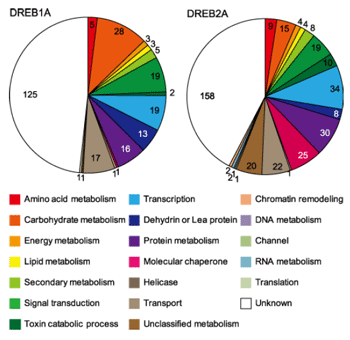
Fig. 2.
Functional categorization of DREB1A and DREB2A-CA downstream genes. Shown are 20 functional categories of DREB1A and DREB2A-CA downstream genes.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources Division
- Classification
-
Administration A
- Term of research
-
FY2009(FY2006~2010)
- Responsible researcher
-
MARUYAMA Kyonoshin ( Biological Resources Division )
YAMAGUCHI-SHINOZAKI Kazuko ( Biological Resources Division )
- ほか
- Publication, etc.
-
Maruyama et al. (2009) Plant Physiol. 150:1972-1980
- Japanese PDF
-
2009_seikajouhou_A4_ja_Part5.pdf87.31 KB