Novel blast resistance genes from a landrace rice variety in Myanmar
Description
The use of broad-spectrum resistance genes is an effective way to achieve durable resistance against rice blast (Pyricularia oryzae Cavara) in rice (Oryza sativa L.).
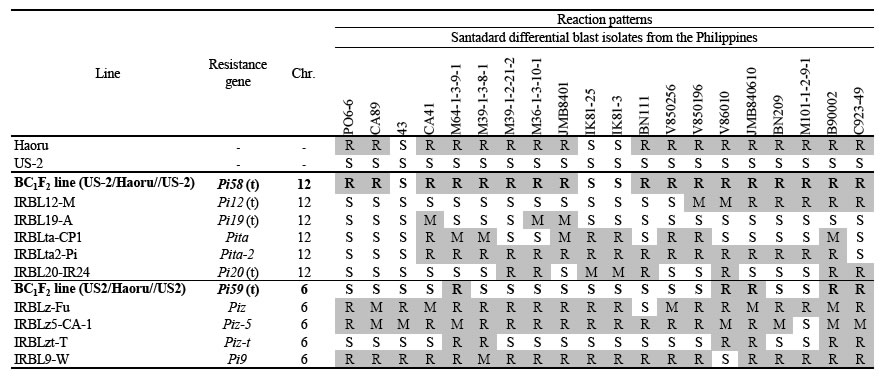
We previously surveyed the diversity of blast resistance in 948 rice varieties and found Haoru (International Rice Research Institute genebank acc. no. IRGC33090), a Myanmar rice landrace with broad-spectrum resistance against blast.
We examined the genetic basis of Haoru’s broad-spectrum resistance using the standard blast differential system consisting of the standard isolates and differential varieties.
For genetic analysis, we used the BC1F1 population and BC1F2 lines derived from crosses of Haoru with a susceptible variety, US-2. Co-segregation analysis of the reaction pattern in the BC1F1 population against the 20 standard isolates suggested that Haoru harbors three resistance genes.
Using bulk-segregant and linkage analysis, we mapped two of the three resistance genes on chromosomes12 and 6, and designated them as Pi58(t) and Pi59(t), respectively.
Pi58(t) and Pi59(t) were differentiated from other reported resistance genes using the standard differential system. The estimated resistance spectrum of Pi58(t) corresponded with that of Haoru, suggesting that Pi58(t) is primarily responsible for Haoru’s broad-spectrum resistance.
In addition, Pi59(t) and the third gene were also proven to be new and useful genetic resources for studying and improving blast resistance in rice.
Figure, table
-
Table 1. Reaction patterns of Haoru and segregation lines harboring new resistance genes to standard differential blast isolates
R: Resistant, M: Moderately resistant, S: Susceptible
-
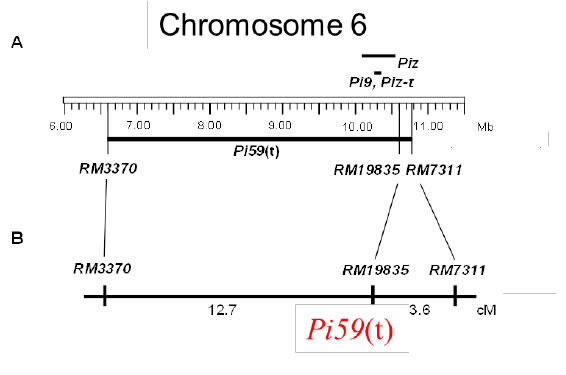
Fig. 1a. Position of resistance gene, Pi59(t), on chromosome 6.
A: Physical map. Position based on the Nipponabare’s genome sequence.
B: Genetic map. Genetic distances between the gene and markers were estimated using the BC1F2 lines of US-2/Haoru//US-2 (n=55). -
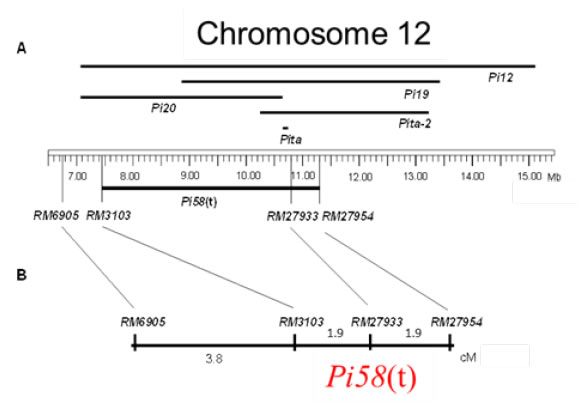
Fig. 1b. Position of resistance gene, Pi58(t), on chromosome 12.
A: Physical map. Position based on the Nipponabare’s genome sequence.
B: Genetic map. Genetic distances between the gene and markers were estimated using the BC1F2 lines of US-2/Haoru//US-2 (n=106).
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources and Post-harvest Division
Japan International Research Center for Agricultural Sciences Tropical Agriculture Research Front
- Classification
-
Administration B
- Research project
- Program name
- Term of research
-
FY 2013(FY2011-FY2015, FY1999-FY2012)
- Responsible researcher
-
Kobayashi Nobuya ( Institute of Crop Science, NARO )
KAKEN Researcher No.: 70252799Koide Yohei ( Research Fellow of the Japan Society for the Promotion of Science )
KAKEN Researcher No.: 70712008Telebanco-Yanoria Mary Jeanie ( Research Fellow of the Japan Society for the Promotion of Science )
Fukuta Yoshimichi ( Tropical Agriculture Research Front )
- ほか
- Publication, etc.
-
https://doi.org/10.1007/s11032-013-9865-5
Koide, Y. et al. (2013) Molecular Breeding, 32: 241-252
- Japanese PDF
-
2013_B06_A3_ja.pdf170.53 KB
2013_B06_A4_ja.pdf263.46 KB
- English PDF
-
2013_B06_A3_en.pdf57.55 KB
2013_B06_A4_en.pdf78.64 KB
- Poster PDF
-
2013_B06_poster.pdf306.5 KB