Genetic variation of blast resistance in rice germplasm from West Africa
Description
Blast disease caused by the fungal pathogen Pyricularia oryzae Cavara is one of the most serious rice diseases worldwide, significantly damaging rice production. Development of cultivars resistant to blast is considered the most effective strategy for protecting the crop, and in West Africa, it is the most economical and effective way of controlling rice blast in the fields of resource-poor farmers. Unfortunately, effective and durable use of blast resistance is limited because the genetic information for resistance to blast disease in rice germplasm from West Africa is quite poor.
To understand the genetic variation and enhance the genetic improvement of rice cultivars in West Africa, we used Simple Sequence Repeat (SSR) markers to investigate the genetic diversity in rice accessions, such as O. sativa including interspecific hybrids (NERICA varieties O. glaberrima and O. barthii), and then evaluated the blast resistance of these accessions. Moreover, we elucidated the relationships between blast resistance and the genome chromosome components of the rice accessions, and discussed the diversity of rice germplasm.
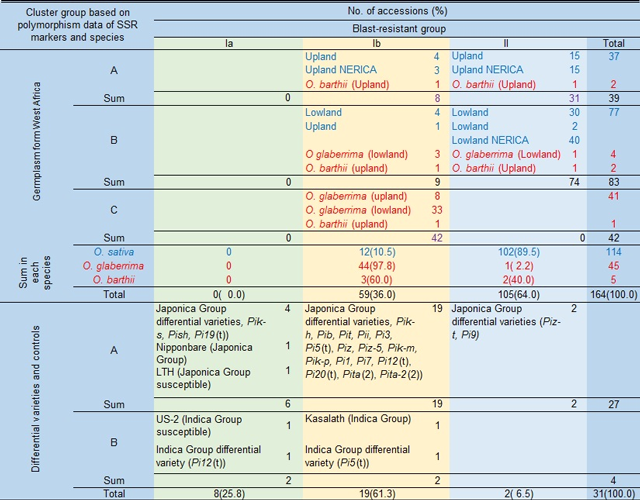
We used 195 rice accessions (Table 1), including O. sativa L. (114 accessions), O. glaberrima (45), O. barthii (5), and differential varieties (DVs) and controls for blast resistance (31), which included two susceptible controls (Lijiangxintuanheigu: LTH and US-2), a Japonica Group cultivar (Nipponbare), and an Indica Group cultivar (Kasalath).
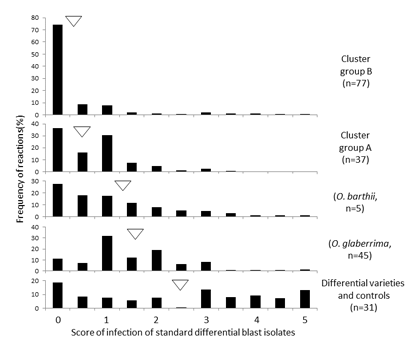
These accessions were classified into three clusters -- the Japonica Group (A), Indica Group (B), and O. glaberrima and O. barthii (C) -- based on the polymorphism data of 61 SSR markers. Moreover, these were classified again into 3 clusters for resistance – low (Ia), middle (Ib) and high (II) -- based on its reactions to 32 international standard differential blast isolates. The resistant group Ia included Nipponbare, LTH, US-2, and five DVs that were lowly resistant. Group Ib had many DVs and accessions from cluster C, and group II were accessions of O. sativa including mainly lowland and upland NERICA cultivars. Many accessions of O. glaberrima were categorized into resistant group Ib, and their resistance were lower than those of lowland and upland cultivars of O. sativa and O. barthii (Fig.1).
These results clearly demonstrated the genetic variation of rice accessions from West Africa, and the relationships among cultivars of O. sativa and O. glaberrima and of wild rice O. barthii. The information will be useful for the genetic improvement of rice cultivars in West Africa.
Figure, table
-
Table 1. Classification of rice accessions from West Africa based on the polymorphism data of DNA markers and the genetic variation in resistance to blast disease
Clusters A and B are corresponded to Japonica Group and Indica Group, respectively, in O. sativa, and cluster C includes O. glaberrima and its wild relative O. barthii.
Group II is highly resistant, and Ib and Ia follow as middle and lowly resistant group, respectively. -
Fig. 1. Resistance of rice accessions from West Africa in each variety group.
The reaction data of rice accessions to 32 standard differential blast isolates are shown in each cultivar group (The data was modified from Odjo et al. 2017).
- Affiliation
-
Japan International Research Center for Agricultural Sciences Tropical Agriculture Research Front
- Research project
- Program name
- Term of research
-
FY2017(2016~2020)
- Responsible researcher
-
Fukuta Yoshimichi ( Tropical Agriculture Research Front )
Yanagihara Seiji ( Biological Resources and Post-harvest Division )
MIERUKA ID: 001780Ojo Theophile ( University of Abomey-Calavi )
Koide Yohei ( Hokkaido University )
KAKEN Researcher No.: 70712008Silue Dress ( AfricaRice )
Kumashiro Takashi ( AfricaRice )
- ほか
- Publication, etc.
-
https://doi.org/10.1270/jsbbs.17051
Odjo T et al. (2017) Breeding Science, 67: 500-508
- Japanese PDF
-
A4 286.79 KB
A3 283.4 KB
- English PDF
-
A4 554.29 KB
A3 319.16 KB
- Poster PDF
-
2017_B01_poster.pdf350.51 KB