QTL for total spikelet number per panicle is detected on chromosome 7 in the genetic background of Indica-type rice cultivar
Description
An Indica-type rice cultivar, IR64, has been widely accepted as high quality rice with good eating quality and high yield in many countries. Total spikelet number per panicle (TSN) is one of the most important traits to increase grain productivity in rice (Oryza sativa L.). To improve yield potential of IR64, a Japanese high-yielding cultivar, Hoshiaoba, was backcrossed to IR64 for three times. We attempted to detect quantitative trait loci (QTL) for TSN by using the developed introgression line. Furthermore, we developed a near isogenic line (NIL) to characterize the effect of the QTL for increasing TSN.
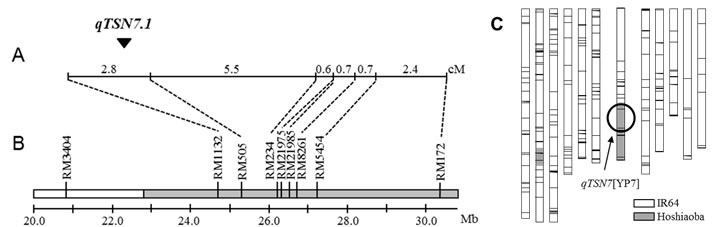
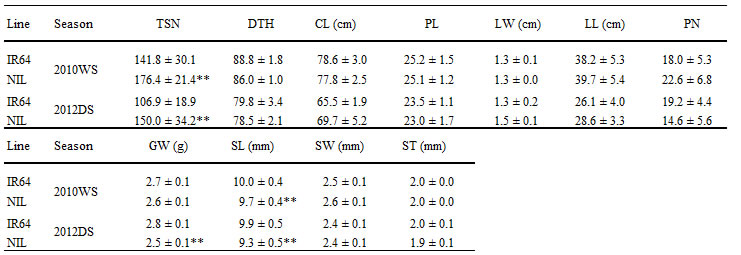
A putative QTL, qTSN7.1,was detected between two markers, RM1132 and RM505, on the long arm of chromosome 7 (Fig. 1A and 1B). For developing NILs, plants which have an introgression in the target chromosomal region of qTSN7.1, were selected from 144 F2 plants derived from a cross between IR64 and its introgression line used for the QTL mapping. Whole-genome survey was conducted using 480 SSR markers distributed throughout the 12 rice chromosomes to generate the graphical genotype of NIL with qTSN7.1 (Fig. 1C). To characterize agronomic traits of NIL, 11 traits were evaluated and compared with those of IR64 (Table 1). NIL showed significantly higher TSN than IR64. In contrast, NIL had significantly shorter SL than IR64. There was no significant difference between IR64 and NIL in other agronomic traits (except GW in 2012DS) across the seasons.
In this study, we succeeded to detect a QTL for TSN, qTSN7.1, using a Japanese high-yielding cultivar, Hoshiaoba, as a donor parent. The developed NIL for TSN with tagged DNA markers would be useful to improve the yield potential of Indica-type rice cultivars through increase in TSN.
Figure, table
-
Fig. 1. Chromosomal location of qTSN7.1 and graphical genotype of NIL with qTSN7.1.
1.Genetic map of the marker on chromosome 7. ▼ ; LOD peak
2.Physical map of the DNA markers. The physical chromosomal position is based on Nipponbare genome sequence.
3.Graphical genotype of the NIL with qTSN7.1. White boxes show the chromosomal segments of IR64, while grey boxes show the chromosomal segments of Hoshiaoba. -
Table 1. Characterization of agronomic traits of IR64 and NIL with qTSN7.1 in the wet season of 2010 and the dry season of 2012.TSN, Total spikelet number; DTH, days to heading; CL, Culm length; PL, Panicle length; LW, Leaf width; LL, Leaf length; PN, Panicle number; GW, 100-grain weight; SL, Seed length; SW, Seed width; ST, Seed thickness. **, significant difference at 1% by t-test.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources and Post-harvest Division
- Classification
-
Administration A
- Research project
- Program name
- Term of research
-
FY 2013 (FY 2005-FY 2013)
- Responsible researcher
-
Kobayashi Nobuya ( Institute of Crop Science, NARO )
KAKEN Researcher No.: 70252799Koide Yohei ( Research Fellow of the Japan Society for the Promotion of Science )
KAKEN Researcher No.: 70712008Fujita Daisuke ( Institute of Crop Science, NARO )
KAKEN Researcher No.: 80721274Tagle Analiza G. ( International Rice Research Institute )
Sasaki Kazuhiro ( University of Tokyo )
KAKEN Researcher No.: 70513688Fukuta Yoshimichi ( Tropical Agriculture Research Front )
Ishimaru Tsutomu ( Biological Resources and Post-harvest Division )
- ほか
- Publication, etc.
-
https://doi.org/10.1007/s10681-013-0882-6
Koide, Y. et al. (2013) Euphytica, 192: 97-106
- Japanese PDF
-
2013_A02_A3_ja.pdf191.66 KB
2013_A02_A4_ja.pdf337.77 KB
- English PDF
-
2013_A02_A3_en.pdf108.96 KB
2013_A02_A4_en.pdf123.49 KB
- Poster PDF
-
2013_A02_poster.pdf267.34 KB