An optimal heat inducible synthetic promoter in plants
Description
Climate change is predicted to adversely affect agriculture. There is a growing need for the development of crops capable of adapting to change. At the molecular level, one of the most discussed environmental conditions is high temperature or heat shock (HS). Many genes have been described that respond to HS stress at the transcriptional level, and their gene products are thought to function in stress tolerance. Heat shock factors (HSFs) bind heat shock response elements (HSEs) with the core sequence to form trimers, thereby regulating downstream gene expression. The importance of the HSF–HSE interaction has previously been suggested in the HS response. However, because plant HSFs are involved not only in HS but also in water deficit stress, development, cell differentiation, and proliferation, it has been suggested that the induction of HS requires a specific combination of HSFs and HSEs.
In this study, to determine representative HS-responsive transcriptional pathways in plants, we analyzed HS-responsible genes and promoters in Arabidopsis, soybean, rice, and maize. We also designed an optimized HS-inducible promoter based on detailed bioinformatics predictions of conserved sequences (cis-acting elements) from HS-inducible promoters of these four plant species for molecular breeding purposes.
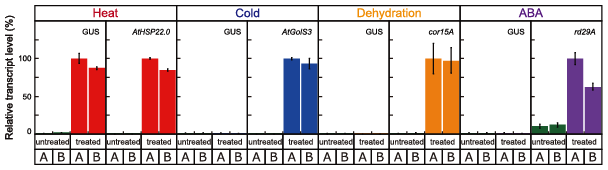
First, to characterize plant HS-responsive genes, a transcriptome analysis of Arabidopsis, soybean, rice, and maize was conducted using microarrays. Second, we used our in-house gene ontology database to annotate the molecular functions of all identified HS-responsive genes. Third, to determine which sequences are conserved in the promoters of HS-inducible genes, we analyzed all hexamer sequences using our promoter research tool. Fourth, based on detailed bioinformatics predictions of conserved sequences, we attempted to design a HS-specific inducible promoter and then performed functional analyses of the designed promoter. The synthetic HSE was placed in an optimal position of promoter, followed by a β-glucuronidase reporter gene (GUS). In our functional analyses, transcript levels of marker genes were significantly higher in cold-, dehydration-, or ABA-treated transgenic plants than in untreated transgenic plants. GUS expression did not increase in each stress-treated transgenic plant; however, its expression increased significantly in HS-treated transgenic plants (Figure 1). We also performed in vivo single-cell gene induction using an infrared laser-evoked gene operator (IR-LEGO) system to observe GUS activity in irradiated cells (Figure 2). These findings demonstrate the utility of our HS-inducible promoter, which we expect to contribute to future molecular breeding of plants adapted to climate change and for the in vivo analysis of gene functions when gene expression is controlled by spatial and/or temporal conditions.
Figure, table
-
Fig. 1. Transcriptional activity of the optimal HS-inducible synthetic promoter. Levels of transcripts for genes encoding β-glucuronidase reporter gene (GUS) and condition-specific markers (heat, AtHSP22.0; cold, AtGols3; dehydration, cor15A; and ABA, rd29A) by quantitative reverse transcription (qRT)-PCR in transgenic plants.
-
Fig. 2. GUS expression induced by infrared laser irradiation of target cells in Arabidopsis lateral root tips. We performed in vivo single-cell gene induction using an infrared laser-evoked gene operator (IR-LEGO) system to observe GUS activity in irradiated cells.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources and Post-harvest Division
- Research project
- Program name
- Term of research
-
FY2016(FY2014~FY2016)
- Responsible researcher
-
Maruyama Kyounoshin ( Biological Resources and Post-harvest Division )
Ogata Takuya ( Biological Resources and Post-harvest Division )
Kanamori Morihito ( Biological Resources and Post-harvest Division )
KAKEN Researcher No.: 30455258Goto Shingo ( National Agriculture and Food Research Organization (NARO) )
Yamamoto Toshiharu ( Gifu University )
KAKEN Researcher No.: 50301784Urawa Hiroko ( Gifu Shotoku Gakuen University )
Iuchi Satoshi ( RIKEN )
KAKEN Researcher No.: 90312256Urano Kaoru ( RIKEN )
Sakurai Tetsuya ( RIKEN )
KAKEN Researcher No.: 90415167Sakakiraba Hitoshi ( RIKEN )
KAKEN Researcher No.: 20242852Shinozaki Kazuo ( RIKEN )
KAKEN Researcher No.: 20124216Shinozaki Kazuko ( University of Tokyo )
ORCID ID0000-0002-0249-8258 - ほか
- Publication, etc.
-
https://doi.org/10.1111/tpj.13420
Maruyama K et al. (2016) Plant J.
- Japanese PDF
-
A4 286.3 KB
A3 214.95 KB
- English PDF
-
A4 527.14 KB
A3 517.22 KB
- Poster PDF
-
Poster 428.21 KB