A new group of nitrogen-fixing endophytic bacteria in rice plants
Description
There are numerous reports on nitrogen-fixing endophytic bacteria that live symbiotically in gramineous plants. We have isolated many bacteria with nitrogen-fixing ability from gramineous pasture grasses and related plants and carried out phylogenetic analyses and inoculation tests on them. However, microorganisms in these ecosystems are not always culturable, although they are viable and their biological activities can be detected. We therefore analyzed communities of nitrogen-fixing endophytes using a culture-independent technique, employing rice as a model plant. RNA-based studies (specifically mRNA) gave information on the activities of specific populations, while DNA-based studies provided information on community structures.
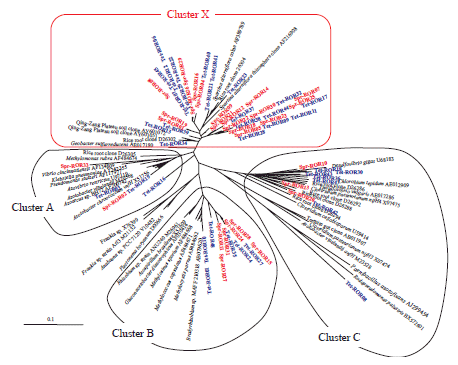
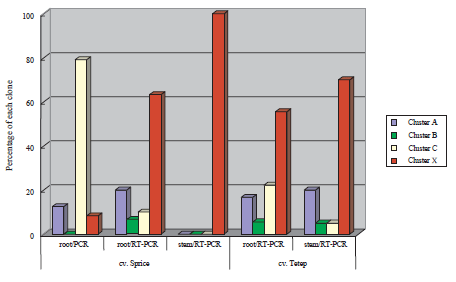
We amplified partial nifH genes by PCR and RT-PCR from DNA and RNA, respectively, extracted from the roots and stems of two rice cultivars (Sprice and Tetep) cultivated in a paddy field, followed by cloning to E. coli and sequencing of the insertions. We carried out a homology search of each NifH amino acid sequence translated from nifH nucleotide sequences, and constructed phylogenetic trees. Numerous NifH sequences without close relatives in known cultured diazotrophs were frequently recovered from the roots and stems of both rice cultivars and designated as “unknown independent Cluster X” (Fig. 1) since they were only distantly related to known diazotrophs. A comparison of roots and stems showed the ratios of Cluster X in stems to be higher than those in roots (Fig. 2). There are several clones belonging to Cluster X identified by PCR that amplify DNA (Fig. 2). Although we very frequently obtained NifH sequences belonging to clusters with Klebsiella (Cluster A), Azospirillum (Cluster B), and Paenibacillus (Cluster C), etc., by the culture method, the frequency of the above sequences recovered by RT-PCR was lower than those using the culture method. These results suggest that the diazotrophs in this new group are truly active and predominant components of nitrogen-fixing endophytes of rice plants and are different from currently known cultured bacteria.
Figure, table
-
Fig. 1.
Phylogenetic analysis (NJ method) of NifH amino acid sequences translated from nifH clones obtained from roots of rice cv. Sprice and cv. Tetep (Spr-RORxx, derived from cv. Sprice; Tet-RORxx, derived from cv. Tetep; xx is the clone number). -
Fig. 2.
Relative abundance of each phylogenetic cluster of NifH translated from nifH clones obtained from roots and stems of rice cv. Sprice and cv. Tetep.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Animal Production and Grassland Division
- Classification
-
Technical A
- Term of research
-
FY2004(FY2003~2005)
- Responsible researcher
-
ANDO Yasuo ( Animal Production and Grassland Division )
ELBELTAGY Adel ( Faculty of Agriculture, Minufiya University )
- ほか
- Publication, etc.
-
Adel Elbeltagy and Yasuo Ando (2005): Phylogenetic analysis of nifH gene sequences from nitrogen-fixing endophytic bacteria associated with the roots of three rice varieties. Journal of Food, Agriculture and Environment, 3, 237-242.
- Japanese PDF
-
2004_11_A3_ja.pdf964.65 KB