Genetic relationship and diversity between and within Mangifera species revealed by AFLP analysis
Description
[Objectives]
Mangifera indica L. (common mango) and its relatives of the genus Mangifera are important crops and can be found throughout tropical Asia. The origin and the center of diversity are thought to be located in Southeast Asia with the greatest diversity occurring in Peninsular Malaysia, Borneo and Sumatra. Currently, classical methods are applied to identify Mangifera species based on phenotypic characteristics such as leaves, fruits and seeds of the plant. However, this method is a slow process due to the long cultivation periods and the evaluation itself, which is easily affected by environmental factors. The objectives were to clarify the effectiveness of AFLP analysis for the identification of Mangifera accessions and to explore the genetic relationship and diversity within and among 4 Mangifera species.
[Results]
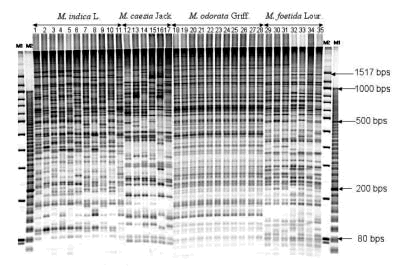
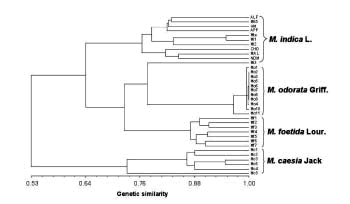
Amplified fragment length polymorphism (AFLP) analysis was utilized to investigate the genetic relationship and diversity among and within 4 Mangifera species. As shown in Fig. 1, we analyzed 35 accessions consisting of 11 accessions (8 cultivars and 3 landraces) of M. indica L., 11 accessions (landraces) of M. odorata Griff., 7 accessions (landraces) of M. foetida Lour., and 6 accessions (landraces) of M. caesia Jack. We carried out AFLP analysis using 8 kinds of primer combinations and 16% polyacrylamide gel. Polymorphic bands among accessions were recorded to clarify diversity and genetic relationship. Eight combinations of primers produced a total of 518 reliable bands in all 35 accessions and a pair of primer combinations yielded 64.8 bands on average. Four-hundred and ninety-nine bands were polymorphic and thus accounted for 96.3% of the total bands. Clustering analysis showed that all 35 accessions were basically classified into 4 groups corresponding to the 4 Mangifera species (Fig. 2). However, the accession in M. indica L., Mi 3, was distantly related to the other M. indica L. accessions and closely related to the M. odorata Griff. group. Although M. odorata Griff. accessions showed very little diversity compared to other Mangifera accessions, this was a suitable method for differentiating between other Mangifera accessions. In the M. caesia Jack accession, Mc 5 was clustered into a group of M. caesia Jack accessions, but this accession was distantly related to the other M. caesia Jack accessions. M. caesia Jack is grouped out from the other species, indicating that its genetic variation is different within the taxa. Our results indicate that the morphological differences and genetic relationship of Mangifera accessions revealed by AFLP analysis are in a good agreement, thus AFLP analysis is an applicable and effective means of conducting taxonomic studies in Mangifera species.
Figure, table
-
Fig. 1. AFLP analysis banding patterns of 35 Mangifera accessions. DNA markers M1 and M2 as well as the E-ACC/M-CTA primer combination were used. -
Fig. 2. A dendrogram of Mangifera accessions displaying the results of AFLP analysis.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources Division
- Classification
-
Technical A
- Term of research
-
FY2002
- Responsible researcher
-
YAMANAKA Naoki ( Biological Resources Division )
HASRAN Masrom ( Malaysian Agricultural Research and Development Institute )
TSUNEMATSU Hiroshi ( Biological Resources Division )
BAN Tomohiro ( Biological Resources Division )
- ほか
- Publication, etc.
-
Yamanaka, N., Hasran, M., Xu, D.H., Tsunematsu, H., Idris, S. and Ban, T. (2005): Genetic relationship and diversity of four Mangifera species revealed through AFLP analysis. Genetic Resources and Crop Evolution (in press).
Hasran, M., Yamanaka, N., Xu, D.H., Tsunematsu, H. and Ban, T. (2003): Genetic relationship of Mangifera accessions revealed by AFLP analysis. Breed. Res., 5, Suppl. 1, 222.
- Japanese PDF
-
2003_07_A3_ja.pdf2.33 MB
- English PDF
-
2003_07_A4_en.pdf78.83 KB