Construction of a molecular linkage map in soybean
Description
[Objectives]
A molecular linkage map covering a large region of the genome with informative DNA markers is very useful for effective and efficient plant selection in breeding programs. It is also essential for identifying and isolating the genes responsible for various quantitative traits. Recently, some soybean linkage maps have been developed with restriction fragment length polymorphism (RFLP) markers based on soybean genomic DNA, and simple sequence repeat (SSR) markers. DNA markers based on polymerase chain reaction (PCR), such as SSR markers, are suitable for the rapid selection of plants in breeding programs. However, DNA markers derived from expressed genes are important for the identification of gene-rich regions as well as quantitative trait loci (QTLs). In addition, since DNA sequences that code for genes are conserved among many plant families, this kind of marker can also be used in other species. Therefore, we constructed a molecular linkage map of soybean by using complementary DNA (cDNA) markers derived from expressed genes in addition to DNA markers based on PCR.
[Results]
The soybean (Glycine max) varieties Misuzudaizu and Moshidou Gong 503 were crossed to develop a segregated population of 190 F2 plants. Based on this population, linkages were calculated among RFLP markers derived from cDNA and genomic DNA, SSR markers, amplified fragment length polymorphism (AFLP) markers, random amplified polymorphic DNA (RAPD) markers, and qualitative trait loci.
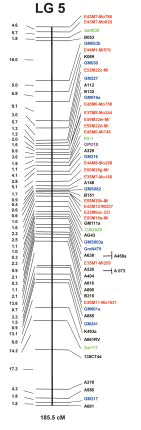
A portion of the constructed soybean molecular linkage map is shown in Fig. 1. This molecular linkage map has 724 markers, including 412 RFLP markers (223 cDNA and 189 genomic DNA), 106 SSR markers, 218 AFLP markers, and one RAPD marker. These markers contain 472 DNA markers developed originally (unpublished data for AFLP markers). This map is the first soybean linkage map having a large number of cDNA markers.
This linkage map also consists of 20 major linkage groups that may correspond to the 20 pairs of soybean chromosomes. The total length of the linkage groups is 3,221 cM of the soybean genome according to the Kosambi function, indicating that this map covers the soybean genome almost completely. This linkage map may thus be used for various aspects of soybean genetic study and breeding.
This study was conducted at Chiba University under partial support from the JIRCAS research project entitled "Comprehensive studies on soybean improvement, production and utilization in South America".
Figure, table
-
Fig. 1. Soybean molecular linkage map of linkage group 5. Distances according to the Kosambi function and markers are shown on the left and right of the bar, respectively. The total length of this linkage group is shown on the bottom. Color separation indicates marker type (i.e. black, RFLP markers derived from genomic DNA; blue, RFLP markers derived from cDNA; green, SSR markers; red, AFLP markers; and purple, RAPD markers).
- Affiliation
-
Chiba University
- Classification
-
Technical A
- Term of research
-
FY2002 (FY1997-2002)
- Responsible researcher
-
HARADA Kyuya ( Chiba University )
YAMANAKA Naoki ( Biological Resources Division )
- ほか
- Publication, etc.
-
Yamanaka, N., Nagamura, N., Tsubokura, Y., Yamamoto, K., Takahashi, R., Kouchi, H., Yano, M., Sasaki, T. and Harada, K. (2000): Quantitative trait locus analysis of flowering time in soybean using a RFLP linkage map. Breeding Science, 50, 109-115.
Yamanaka, N., Ninomiya, S., Hoshi, M., Tsubokura, Y., Yano, M., Nagamura, Y., Sasaki, T. and Harada, K. (2001): An informative linkage map of soybean reveals QTLs for flowering time, leaflet morphology and regions of segregation distortion. DNA Research, 8, 61-72.
Hossain, K.G., Kawai, H., Hayashi, M., Hoshi, M., Yamanaka, N. and Harada, K. (2000): Characterization and identification of (CT) nmicrosatellites in soybean using sheared genomic libraries. DNA Research, 7, 103-110.
Toda, K., Yang, D., Yamanaka, N., Watanabe, S., Harada, K. and Takahashi, R. (2002): A single-base deletion in soybean flavonoid 3'-hydroxylase gene is associated with gray pubescence color. Plant Molecular Biology, 50, 187-196.
Nishimura, R., Hayashi, M., Wu, G. J., Kouchi, H., Imaizumi-Anraku, H., Murakami, Y., Kawasaki, S., Akao, S., Ohmori, M., Nagasawa, M., Harada, K. and Kawaguchi, M. (2002): HAR1 mediates systemic regulation of symbiotic organ development. Nature, 420, 426-429.
- Japanese PDF
-
2002_15_A3_ja.pdf1.13 MB
- English PDF
-
2002_15_A4_en.pdf53.24 KB