Simple and rapid method for the detection of lipoxygenase isozymes in soybean seeds
Description
[Objectives]
In South American countries, the soybean has been used mainly as a source of edible oil and in livestock feed, while direct food consumption, as is common in Asia, is very limited. Soybeans used for the purposes of food consumption, however, possess great potential for improving the nutrition and health of the local population and for generating income. The limitation on direct consumption in this region is associated with undesirable flavors in soybean food products. Since lipoxygenases have been found to be responsible for grassy-beany flavors, it would therefore be highly desirable to remove these enzymes in locally-adapted soybean cultivars. Through the use of mutants lacking lipoxygenases, L-1, L-2, and L-3, the genetic approach towards breeding triple mutant soybean lacking all isozymes has been employed, aiming to reduce undesirable flavors. In addition, a lipoxygenase isozyme detection method suitable for routine screening is becoming an increasingly utilized approach. The presence or absence of the three lipoxygenase isozymes can be readily and accurately determined by spectrophotometric methods, based on the different bleaching abilities of L-1, L-2 and L-3 isozymes in contact with methylene blue and s-carotene. However, this method is time-consuming, as different seed samples are necessary for the analysis of each isozyme. The Division's attempt was based upon this above method and was carried out to develop a simpler and more rapid visual method for the detection of individual lipoxygenase isozymes in soybean seeds.
[Results]
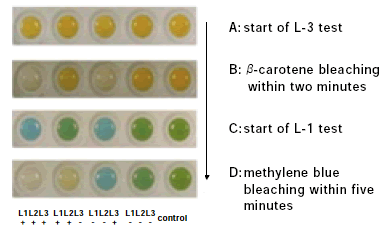
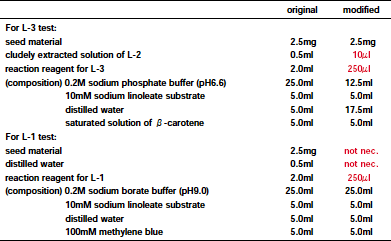
After several attempts, we succeeded in finding a method by which identification of the lipoxygenase isozymes was possible using only a small amount of milled soybean seed samples (Table 1). The new method also enabled the identification of two isozymes, L-1 and L-3, by using the same seed sample and adding the reaction reagent for the L-1 isozyme (Fig. 1). Within five minutes, it was possible to visually determine the individual lipoxygenase phenotypes as follows: colorless - normal soybean having all isozymes; yellow- single mutant lacking L-3; blue - double mutant lacking L-1 and L-2; green - triple mutant lacking all isozymes (Figs. 1, 2). In crosses between triple mutant soybeans and normal soybeans, the other test which is used to identify the L-2 isozyme may be omitted, since the Lx1 locus is closely linked to the Lx2 locus. By using this simplified new method, the genetic variation of these isozymes contained in South American soybean cultivars and genetic lines can be efficiently researched.
Figure, table
-
Fig. 1. Visual detection of individual lipoxygenase phenotypes of soybean seeds derived from crosses between triple mutant lacking all isozymes and normal type. -
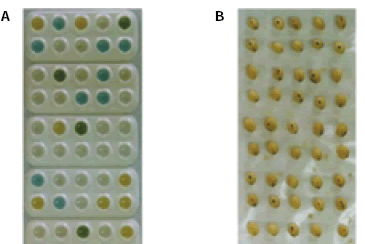
Fig. 2. Analyses of F2 seeds derived from the cross between triple mutant lacking all isozymes and normal type. A: result of analyses, B: seeds corresponding to analyses. -
Table 1. Comparison of original (Suda et al., 1995) and modified methods to identify lipoxygenase isozymes in each soybean seed.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources Division
- Classification
-
Technical A
- Term of research
-
FY2001 (FY1998-2001)
- Responsible researcher
-
KIKUCHI Akio ( Biological Resources Division )
- ほか
- Japanese PDF
-
2001_03_A3_ja.pdf740.57 KB
- English PDF
-
2001_03_A4_en.pdf63.05 KB