Development of wild soybean chromosome segment substitution lines for genetic studies of important traits
Description
Wild soybean (Glycine soja Sieb. & Zucc.) is believed to be the progenitor of cultivated soybean (G. max (L.) Merr.). It has been revealed that wild soybean has higher genetic diversity than cultivated soybean, presenting a potential genetic resource pool for improvement of the latter. However, the inheritance mode of many important agronomic traits, such as seed quality, grain yield, and tolerance to environmental stresses, is extremely complex, and their performances are greatly affected by the genetic background and growth environment, making it difficult to identify the useful genes possessed by wild soybean. In this study, we created wild soybean chromosome segment substitution lines (CSSLs), in which the cultivated soybean chromosomes were replaced by only one or a few small chromosome segments of wild soybean, and thus these lines have similar genetic background but differ only in the small wild soybean chromosome region(s). We employed the CSSLs for genetic analysis of some important agronomic traits in soybean and demonstrated that the CSSLs might accelerate the identification of useful genes in wild soybean.
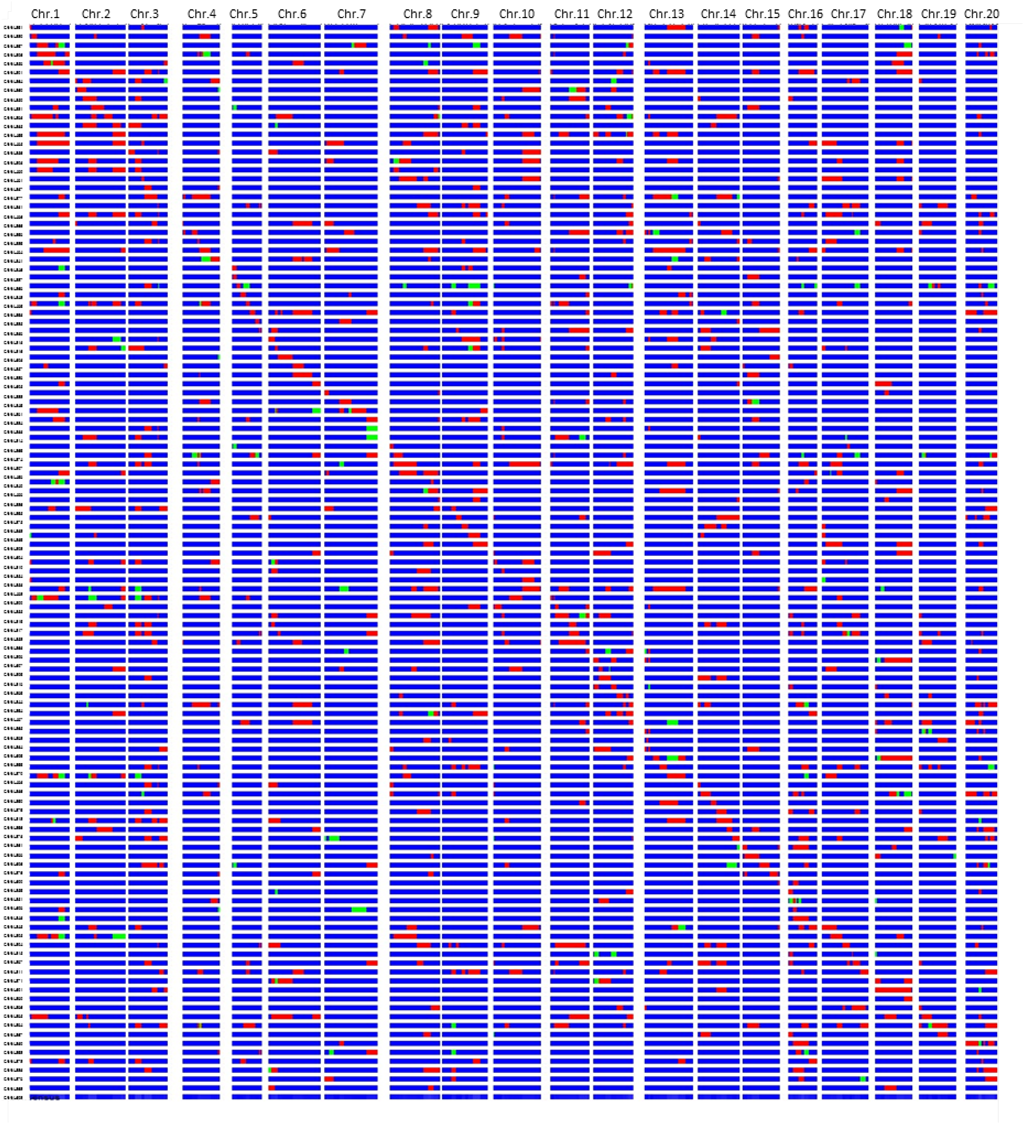
A total of 120 BC3F5 CSSLs were developed from a cross between the cultivated soybean variety ‘Jackson’ and the wild soybean accession ‘JWS156-1.’ These lines were created by backcrossing of {[((‘Jackson’ × ‘JWS156-1’) × ‘Jackson’) × ‘Jackson’] × ‘Jackson’} and five successive generations of self-pollination without any selection. A total of 235 SSR soybean markers from all 20 chromosomes were used for genotyping the 120 CSSLs. The proportion of the recurrent parent ‘Jackson’ alleles in each CSSL ranged from 80.3% to 99.2% with an average of 92.9 ± 4.0%, which corresponded to the expected value, 93.8% (Fig. 1).
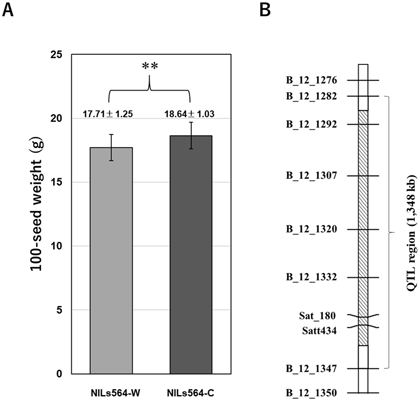
To identify the quantitative trait locus (QTL) of seed weight, which is one of the most important traits that control soybean yield, the 120 CSSLs were cultivated in field conditions over three years and the seed weight trait was measured for each CSSL. QTL analysis detected nine QTLs (qSW8.1, qSW9.1, qSW12.1, qSW13.1, qSW14.1, qSW16.1, qSW17.1, qSW17.2, and qSW20.1) on eight chromosomes. Of these, qSW12.1 was detected over the three successive years on a region of 1,348 kb in chromosome 12 as a novel, stable, and major seed weight QTL (Fig. 2).
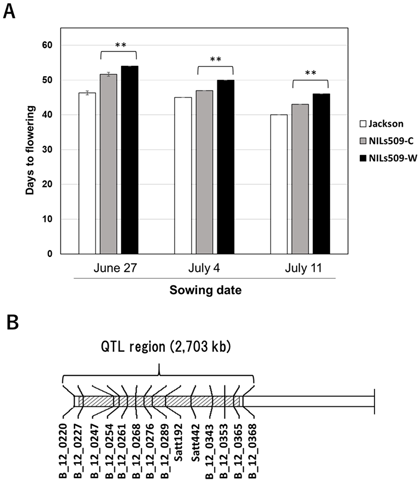
In order to identify new flowering time QTLs, we evaluated the 120 CSSLs over two years under field conditions. Four QTLs (qFT07.1, qFT12.1, qFT12.2, and qFT19.1) were detected on three chromosomes. Of these, qFT12.1 showed the highest effect, accounting for 36.37–38.27% of the total phenotypic variation over two years. qFT12.1 may be a new flowering time gene locus in soybean (Fig. 3).
In our study, a BC3F5 CSSL population has been developed. Since some CSSLs still has wild soybean segments or heterozygous regions other than the target wild soybean substitution segment, further backcrossing and DNA marker selection are needed for eliminating such redundant wild segments in the CSSLs. Nevertheless, the developed CSSL population has potential for mining useful genes in wild soybean.
Figure, table
-
Fig. 1. Graphical genotypes of the wild soybean ‘JWS156-1’ chromosome segment substitution lines (CSSLs) in the cultivated soybean variety ‘Jackson’ background.
Red: ‘JWS156-1’ homozygous, blue: ‘Jackson’ homozygous, green: heterozygous. This figure is modified from Liu D et al. (2018a) (Copyright: Japanese Society of Breeding). -
Fig. 2. Seed weight QTL qSW12.1 detected in the wild soybean CSSLs population.
(A) The allele effect of qSW12.1 in near-isogenic lines NILs564-C (‘Jackson’ genotype) and NILs564-W (‘JWS156-1’ genotype). Data are shown as means ± SD (standard deviation). **: P < 0.01. (B) qSW12.1 was delimited in a 1,348-kb interval on chromosome 12. This figure is modified from Liu D et al. (2018a) (Copyright: Japanese Society of Breeding). -
Fig. 3. Flowering time QTL qFT12.1 detected in the wild soybean CSSL populationa
(A) Allelic effects of qFT12.1 in near-isogenic lines (NILs) sown in the field on different dates in 2017. NILs509-C: ‘Jackson’ genotype, NILs509-W: ‘JWS156-1’ genotype. Data are shown as means ± SD (standard deviation). **: P < 0.01. (B) qFT12.1 was delimited in a 2,703-kb interval on chromosome 12. This figure is modified from Liu D et al. (2018b) (Copyright: Springer Science + Business Media B.V.).
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources and Post-harvest Division
- Research project
- Program name
- Term of research
-
FY2018(FY2016-FY2018)
- Responsible researcher
-
Xu Donghe ( Biological Resources and Post-harvest Division )
Fujita Yasunari ( Biological Resources and Post-harvest Division )
Liu Dequan ( Biological Resources and Post-harvest Division )
- ほか
- Publication, etc.
-
https://doi.org/10.1270/jsbbs.17127
Liu D et al. (2018) Breeding Science, 68(4):442-448
https://doi.org/10.1007/s11032-018-0808-zLiu D et al. (2018) Molecular Breeding, 38:45
- Japanese PDF
-
A4531.77 KB
A3260.22 KB
- English PDF
-
A4413.01 KB
A3307.59 KB
- Poster PDF
-
2018_B04_poster.pdf617.04 KB