A flexible high-throughput marker system to distinguish African rice (Oryza glaberrima ) from Asian rice (Oryza sativa )
Description
The African rice Oryza glaberrima is an important reservoir of genes for abiotic stress tolerance. To discover such tolerance genes and to exploit them in rice improvement, a flexible, high-throughput marker system is needed. Single nucleotide polymorphism (SNP) sites, where the genome sequence of two or more individuals differs by a single base, are increasingly becoming the marker of choice. Although the number of discovered SNPs has increased significantly over the past few years, most of these efforts have focused on variations within the Asian rice Oryza sativa. The aim of the present study was to detect a set of SNPs that differentiate between O. sativa and O. glaberrima, and to use a representative subset to develop a high-throughput PCR-based genotyping panel.
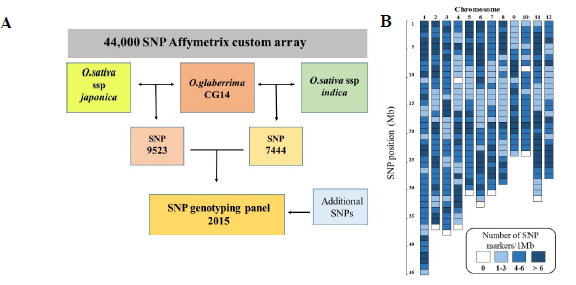
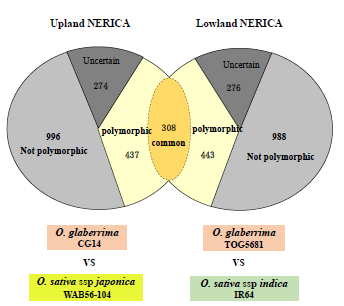
A genome-wide 44,000 SNP genotyping array identified a set of 9523 SNPs polymorphic between O. glaberrima and O. sativa subspecies indica, and 7444 SNPs between O. glaberrima and O. sativa subspecies japonica (Figure 1A). From the above set, a subset of 1540 SNPs was selected in collaboration with partners within the Generation Challenge Program (GCP) for conversion into PCR-based markers, using the KASP (competitive-allele PCR) technology. A final set of 2015 SNPs were successfully converted to KASP markers, which are evenly distributed in the genome, with the exception of small gaps in chromosomes 4 and 10 (Figure 1B). The panel was validated in the ‘New Rice for Africa’ (NERICA) parents. Of the 2015 markers tested, 745 markers were polymorphs between CG14 and WAB56-104 (upland NERICA parents) and 752 between TOG5681 and IR64 (lowland NERICA parents) (Figure 2). Several subsets of these markers have been used successfully to map O. glaberrima introgressions in NERICA rice varieties and interspecific breeding lines.
This new genotyping panel is a cost-effective, gel-free genotyping platform that allows maximum flexibility for ‘pick-and-choose’ markers according to individual breeder’s needs. Presently, it is fully outsourced to service companies that can perform all steps, from DNA extraction to genotyping, which would extend the capacity of low-resource laboratories to perform molecular breeding using local rice varieties. In addition, the information for each SNP is publicly available allowing the rice breeding community to complement the set with their own subset of markers. JIRCAS is expanding its collaborative rice breeding network with applications ranging from parental surveys, development of QTL mapping populations, and marker-assisted introgressions of major stress tolerance genes like OsPSTOL1.
Figure, table
-
Fig. 1. Flow chart indicating the selection of polymorphic SNPs between O. glaberrima vs O. sativa ssp japonica and indica for conversion into PCR-based markers (A). Distribution of SNP markers along the rice genome. Color represents the number of markers per 1 Mb (B).
-
Fig. 2. Distribution of polymorphic SNP markers for crosses of O. glaberrima vs O. sativa. There are 745 and 751 polymorphic markers for upland and lowland NERICA varieties, respectively.
- Affiliation
-
Japan International Research Center for Agricultural Sciences Crop, Livestock and Environment Division
- Classification
-
Administration A
- Research project
- Program name
- Term of research
-
FY 2014 (FY 2011-FY 2015)
- Responsible researcher
-
Wissuwa Matthias ( Crop, Livestock and Environment Division )
KAKEN Researcher No.: 90442722Tanaka Juan ( Crop, Livestock and Environment Division )
Lorieux Mathias ( International Center for Tropical Agriculture )
He Chunlin ( International Maize and Wheat Improvement Center )
McCouch Susan ( Cornell University )
Thomson Michael J. ( International Rice Research Institute )
- ほか
- Publication, etc.
-
https://doi.org/10.1007/s10681-014-1183-4
J. Pariasca-Tanaka et al. (2014) Euphytica.
- Japanese PDF
-
2014_B02_A3_ja.pdf251.75 KB
2014_B02_A4_ja.pdf353.46 KB
- English PDF
-
2014_B02_A3_en.pdf234.21 KB
2014_B02_A4_en.pdf251.87 KB
- Poster PDF
-
2014_B02_poster.pdf326.31 KB