Pathogenic variation of soybean rust in South America
Description
Soybean [Glycine max (L.) Merrill] is an economically important crop and is a valuable source of oil and protein worldwide as well as of food products traditional to the Orient. South American countries are the largest soybean producers in the world, with production centered in Brazil, Argentina, and Paraguay. Soybean rust, caused by Phakopsora pachyrhizi Sydow & P. Sydow, is one of the most destructive and economically important diseases of soybean. Understanding the pathogenicity of indigenous fungal populations is useful for identifying resistant plant genotypes and targeting effective cultivars against certain populations. The objective of this study was to investigate pathogenicity of P. pachyrhizi infecting soybean in the 3 South American countries in 2007–2010, and to compare the rust pathogenicity geographically and temporally.
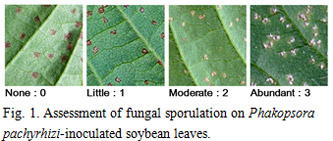
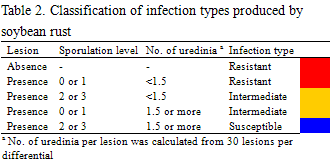
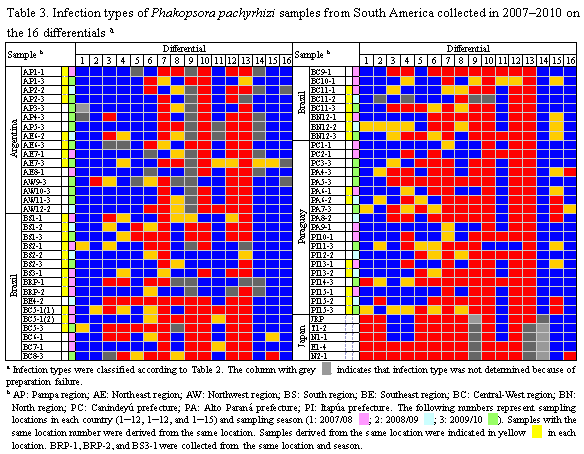
Soybean rust samples were collected in Argentina, Brazil, and Paraguay in the 3 cropping seasons of 2007/2008, 2008/2009, and 2009/2010. For comparative analysis of soybean rust pathogenicity between South America and Japan, rust samples were collected in Japan in 2007 and 2008. A total of 16 soybean genotypes including cultivars and lines (Table 1) were selected as a differential set to test soybean rust populations from South American countries. Nine differentials were reported to carry Rpp (resistance to P. pachyrhizi) genes (Table 1): Rpp1 in Plant Introduction (PI) 200492, PI 368039, PI 587880A, and PI 587886; Rpp2 in PI 230970 and PI 417125; Rpp3 in PI 462312; Rpp4 in PI 459025; and Rpp5 in Shiranui. Sixteen soybean differentials were grown at 24°C with a 14 h photoperiod under rust-free conditions in a growth chamber and inoculated with P. pachyrhizi urediniospore suspension using a paintbrush or a glass atomizer. Two weeks after inoculation, lesion appearance [presence (+) or absence (−) of lesions] and sporulation level (SL) on the differential set were determined macroscopically. SL was rated using a 0–3 scale: 0, none; 1, little; 2, moderate; 3, abundant (Fig. 1). A few soybean leaves for each differential were detached from the inoculated plants, and the number of uredinia per lesion (NoU) formed on the abaxial side of the leaves were counted under a stereomicroscope. The NoU was calculated from 30 lesions per genotype. Data for the 3 parameters, (i) presence (+) or absence (−) of lesions, (ii) SL, and (iii) NoU, were collected for all rust populations and converted into infection types caused by the rust populations (Table 2). Infection types without lesions (immune) and with lesions showing SL 0 or 1 and NoU <1.5 were classified as resistant (R) indicated in red, whereas those with lesions showing SL 2 or 3 and NoU ≥1.5 were classified as susceptible (S) indicated in blue. When lesions with SL 2 or 3 and NoU <1.5 or SL 0 or 1 and NoU ≥1.5 were observed, the infection types were classified as intermediate (IM) indicated in yellow.
Fifty-nine rust samples from Argentina, Brazil, and Paraguay in 3 cropping seasons of 2007–2010 were evaluated for pathogenicity using 16 soybean differentials (Table 3). In the South American populations analyzed, only 2 pairs of populations yielded identical pathogenicity profiles in the 16 differentials: BE4-2 and PA5-3 from Brazil and Paraguay, respectively, and PC1-1 and PA9-1 from Paraguay, indicating substantial pathogenic variation in the rust populations. Each of the rust samples with identical pathogenicity profiles was collected from different locations, suggesting no association between pathogenicity and geographical origins of the samples. Comparative analysis of 59 South American and 5 Japanese samples revealed that pathogenic differences were not only detected within South America but also distinct between the P. pachyrhizi populations from South America and Japan. In addition, pathogenic differences were observed among South American P. pachyrhizi populations with the same geographical origin but different temporal origins. Thus, yearly changes in rust pathogenicity were detected during the sampling period. The differentials containing resistance genes Rpp1, Rpp2, Rpp3, and Rpp4, except for PI 587880A, displayed resistant reaction to only 1.8%–14%, 24%–28%, 22%, and 36% of South American P. pachyrhizi populations, respectively. In contrast, PI 587880A (Rpp1), Shiranui(Rpp5), and 3 Rpp-unknown differentials, PI 587855, PI 587905, and PI 594767A showed resistant reaction to 78%–96% of all populations. This study demonstrated pathogenic diversity of P. pachyrhizi populations in South America and that the known Rpp genes other than Rpp1 in PI 587880A and Rpp5 have been less effective against recent pathogen populations in the countries studied.
Figure, table
- Affiliation
-
Japan International Research Center for Agricultural Sciences Biological Resources and Post-harvest Division
- Classification
-
Administration A
- Research project
- Program name
- Term of research
-
FY 2012 (FY 2011-FY 2015)
- Responsible researcher
-
Akamatsu Hajime ( Biological Resources and Post-harvest Division )
Yamanaka Naoki ( Biological Resources and Post-harvest Division )
MIERUKA ID: 001770Yamaoka Yuuichi ( University of Tsukuba )
KAKEN Researcher No.: 00220236Ivancovich Antonio Juan Gerardo ( Instituto Nacional de Tecnologia Agropecuaria )
Soares Rafael Moreira ( Brazilian Agricultural Research Corporation )
Morel Wilfrido ( The Paraguayan Institute of Agrarian Technology )
Bogado Alicia Noelia ( The Paraguayan Institute of Agrarian Technology )
- ほか
- Publication, etc.
-
Akamatsu et al. (2013) Journal of General Plant Pathology 79(1):28-40.
- Japanese PDF
-
2012_09_A4_ja.pdf168.24 KB
- English PDF
-
2012_09_A4_en.pdf269.6 KB